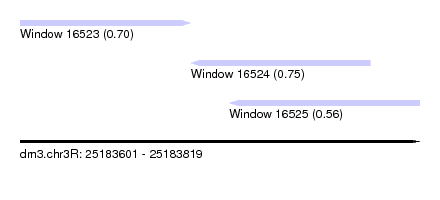
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,183,601 – 25,183,819 |
| Length | 218 |
| Max. P | 0.749822 |

| Location | 25,183,601 – 25,183,694 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 71.11 |
| Shannon entropy | 0.59420 |
| G+C content | 0.41476 |
| Mean single sequence MFE | -20.87 |
| Consensus MFE | -9.33 |
| Energy contribution | -9.02 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.704529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25183601 93 + 27905053 UUAAUAAUUCACUUUGCGCAGUCAAUUUGGCCAAAUCGCUUUGGCUAUUCGAGAUGGCGAAAGUCCGGCC-AUUUUUUUUUUUUUUUGUCUCUU------ ..........(((((.(((.(((....((((((((....)))))))).....))).))))))))......-.......................------ ( -18.60, z-score = -0.49, R) >droSim1.chr3R 24858533 85 + 27517382 UUAAUAAUUCACUUUGCGCAGUCAAUUUGGCCAAAUCGCUUUGGCUAUUCGAGAUGGCGAAAGUCCGGCC-AUUUUUUUGUCUCUU-------------- .................((((..((..(((((.....(((((.((((((...)))))).)))))..))))-)..)).)))).....-------------- ( -20.00, z-score = -0.90, R) >droSec1.super_4 4043756 85 + 6179234 UUAAUAAUUCACUUUGCGCAGUCAAUUUGGCCAAAUCGCUUUGGCUAUUCGAGAUGGCGAAAGUCCGGCC-AUUUUUUUGUCUCUU-------------- .................((((..((..(((((.....(((((.((((((...)))))).)))))..))))-)..)).)))).....-------------- ( -20.00, z-score = -0.90, R) >droYak2.chr3R 7528905 87 - 28832112 UUAAUAAUUCACUUUGCGCAGUCAAUUUGGCCAAAUCGCUUUGGCUAUUCGAGAGAGCCUUAUUUUUUUUUAUUUUGUUGUCUGUUG------------- .................((((.((((.((((((((....))))))))(((....)))...................)))).))))..------------- ( -17.50, z-score = -1.34, R) >droEre2.scaffold_4820 7630116 93 - 10470090 UUAAUAAUUCACUUUGCGCAGUCAAUUUGGCCAAAUCGCUUUGGCUAUUCGAGAGAGCAAAAGUCCGGGC-CUUUUUUUUUGUUUUCGUCUGUU------ .................((((.(....((((((((....)))))))).....(((((((((((.......-.....)))))))))))).)))).------ ( -22.50, z-score = -1.20, R) >droAna3.scaffold_12911 4875783 85 + 5364042 UUAAUAAUUCACUUUGCGCAGUCAAUUUGGCCAAAUCGCUUUGGCUAUUCG-----GCCUAGGCCUUUCUCUUUUUUUAC-GUAUUCUGUG--------- ................(((((......((((((((....))))))))...(-----((....)))...............-.....)))))--------- ( -18.60, z-score = -2.17, R) >dp4.chr2 2040236 96 + 30794189 UUAAUAAUUCACUUUGCGCAGUCAAUUUGGUCAAAUCGCCUUGGCCAUUUCAG---GCCUAGCCUGGCCUGCCU-UUUACUGUGUGCUGUCCCGUCCGGC ...............(((((((.....(((((((......)))))))...(((---(((......))))))...-...)))))))((((.......)))) ( -29.50, z-score = -2.19, R) >droPer1.super_7 2207934 96 + 4445127 UUAAUAAUUCACUUUGCGCAGUCAAUUUGGUCAAAUCGCCUUGGCCAUUUCAG---GCCUAGCCUGGCCUGCCU-CUUACUGUGUGCUGUCCCGUCCGGC ...............(((((((.....(((((((......)))))))...(((---(((......))))))...-...)))))))((((.......)))) ( -29.50, z-score = -2.24, R) >droWil1.scaffold_181089 7956410 84 + 12369635 UUAAUAAUUCACUUUGCGCAGUCAAUUUGGCCAAAUCGC-UUGGCUAUU-CAG---GCUGA--CCGGCUUGCUUGCUUGCUUUCAUUUCUU--------- .................((((.(((..(((((((.....-)))))))..-(((---(((..--..)))))).))).))))...........--------- ( -19.50, z-score = -0.61, R) >droGri2.scaffold_14624 3427298 84 + 4233967 UUAAUAAUUCACUUUACGCAGUCAAUUUAGCCAAAUCGCUU-GGCUAUUCG-----GCC--GGCCUUUUUCAUUUUUUGUUAUAUGCUUUAU-------- .................(((..(((..(((((((.....))-)))))...(-----(..--..))...........))).....))).....-------- ( -13.00, z-score = -0.52, R) >consensus UUAAUAAUUCACUUUGCGCAGUCAAUUUGGCCAAAUCGCUUUGGCUAUUCGAG___GCCAAAGCCCGGCU_AUUUUUUUCUGUCUUCU_U__________ .................((........(((((((......))))))).........)).......................................... ( -9.33 = -9.02 + -0.31)
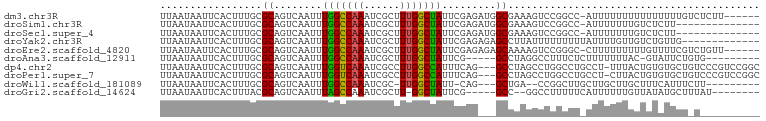
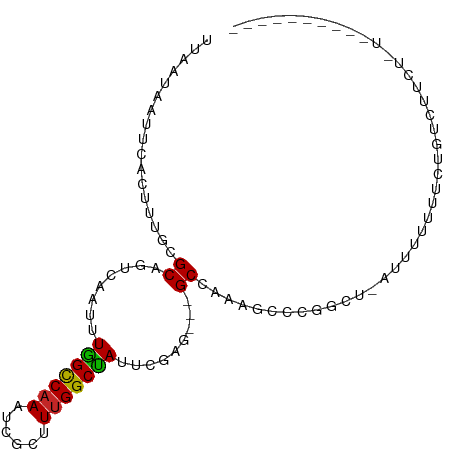
| Location | 25,183,694 – 25,183,792 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 85.40 |
| Shannon entropy | 0.28236 |
| G+C content | 0.36883 |
| Mean single sequence MFE | -20.16 |
| Consensus MFE | -14.99 |
| Energy contribution | -15.33 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25183694 98 - 27905053 --ACGAAUGACAGUUAAGUGACAUUUAACGGUUAAUAAGCCUGACAGAUUUACAACUGUCAGCAGCGUUUAAAAAUAAUGAACAUCAAC--AUCAGCGACGG---------------- --.((..((((.(((((((...))))))).))))....(((((((((........)))))))....(((((.......)))))......--....))..)).---------------- ( -19.70, z-score = -1.50, R) >droSim1.chr3R 24858618 98 - 27517382 --ACGAAUGACAGUUAAGUGACAUUUAACGGUUAAUAAGCCUGACAGAUUUACAACUGUCAGCGGCGUUUAAAAAUAAUGAACAUCAAC--AUCAGCGACGG---------------- --.((..((((.(((((((...))))))).))))....(((((((((........))))))).(..(((((.......)))))..)...--....))..)).---------------- ( -20.30, z-score = -1.41, R) >droSec1.super_4 4043841 98 - 6179234 --ACGAAUGACAGUUAAGUGACAUUUAACGGUUAAUAAGCCUGACAGAUUUACAACUGUCAGCGACGUUUAAAAAUAAUGAACAUCAAC--AUCAGCGACGG---------------- --.((..((((.(((((((...))))))).))))....(((((((((........))))))).((.(((((.......))))).))...--....))..)).---------------- ( -21.00, z-score = -1.99, R) >droYak2.chr3R 7528992 98 + 28832112 --ACGAAUGACAGUUAAGUGACGUUUAACGGUUAAUAAGCCUGACAGAUUUACAACUGUCAGCAACGUUUAAAAAUAAUGAACAUCAAC--AUCAGCGACGG---------------- --.((..((((.((((((.....)))))).))))....(((((((((........)))))))....(((((.......)))))......--....))..)).---------------- ( -19.30, z-score = -1.10, R) >droEre2.scaffold_4820 7630209 98 + 10470090 --GCGAAUGACAGUUAAGUGACGUUUAACGGUUAAUAAGCCUGACAGAUUUACAACUGUCAGCAACGUUUAAAAAUAAUGAACAUCAAC--AUCAGCGACGG---------------- --((..(((...(((....)))(((((...(((..((((((((((((........)))))))....)))))..)))..))))).....)--))..)).....---------------- ( -19.50, z-score = -0.99, R) >droAna3.scaffold_12911 4875868 93 - 5364042 --ACGAAUGACAGUUAAGUGACAUUUAACGGUUAAUAAGCCUGACAGAUUUACAACUGUCAGCGACGUUUAAAAAUAAUGAACAUCAACUGACAG----------------------- --..(((((.((......)).)))))..(((((.....((.((((((........))))))))((.(((((.......))))).)))))))....----------------------- ( -19.20, z-score = -2.14, R) >dp4.chr2 2040332 100 - 30794189 --ACGAAUGACACUUAAGUGACAUUUAACGGUUAAUAAGUCUGACAGAUUUACAACUGUCAGCGACGUUUAAAAAUAAUGAACAGCAACUGACAGGCAACGA---------------- --..(((((.(((....))).)))))..(((((.....(.(((((((........))))))))...(((((.......)))))...)))))...(....)..---------------- ( -22.60, z-score = -2.41, R) >droPer1.super_7 2208030 100 - 4445127 --ACGAAUGACACUUAAGUGACAUUUAACGGUUAAUAAGUCUGACAGAUUUACAACUGUCAGCGACGUUUAAAAAUAAUGAACAGCAACUGACAGGCAACGA---------------- --..(((((.(((....))).)))))..(((((.....(.(((((((........))))))))...(((((.......)))))...)))))...(....)..---------------- ( -22.60, z-score = -2.41, R) >droWil1.scaffold_181089 7956494 101 - 12369635 ACUCAGAUGACAGUUAAGUGACAUUUAACGGUUAAUAAGCCUGACAGAUUUACAACUGUCAGCGGCUUUUAAAAAUAACGAACCUCAAG-GACAAAGGACGA---------------- ..((...((((.(((((((...))))))).))))..(((((((((((........))))))..)))))...........)).((....)-)...........---------------- ( -19.60, z-score = -1.09, R) >droGri2.scaffold_14624 3427382 117 - 4233967 -AAAGCAUGACAGUUAAGUGACAUUUAACGGUUAAUAAGCCUGACAGAUUUACAACUGUCAACGACGUUUAAAAAUAAUGAACAACGAGGACCAAAUAAAAAAGAAAUAUAUAGAAAU -...........(((((((...)))))))((((........((((((........)))))).((..(((((.......)))))..))..))))......................... ( -17.80, z-score = -2.19, R) >consensus __ACGAAUGACAGUUAAGUGACAUUUAACGGUUAAUAAGCCUGACAGAUUUACAACUGUCAGCGACGUUUAAAAAUAAUGAACAUCAAC__ACAAGCGACGG________________ .......((((.(((((((...))))))).))))....(.(((((((........))))))))...(((((.......)))))................................... (-14.99 = -15.33 + 0.34)

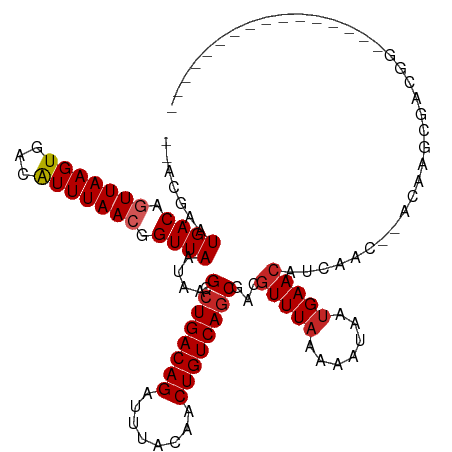
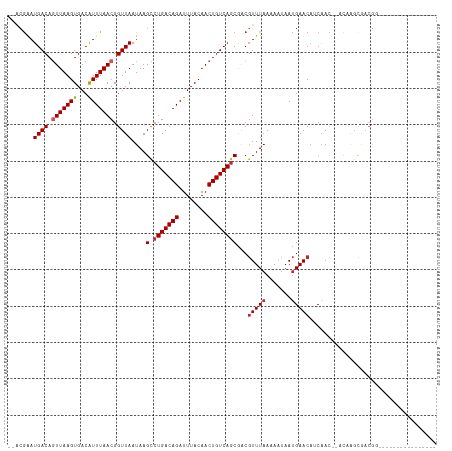
| Location | 25,183,715 – 25,183,819 |
|---|---|
| Length | 104 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.15 |
| Shannon entropy | 0.35747 |
| G+C content | 0.39417 |
| Mean single sequence MFE | -27.22 |
| Consensus MFE | -17.13 |
| Energy contribution | -16.51 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.557344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25183715 104 - 27905053 -----------GCGGCU-CGCGUCGAAUUG--UUAUGGCCAA--CGAAUGACAGUUAAGUGACAUUUAACGGUUAAUAAGCCUGACAGAUUUACAACUGUCAGCAGCGUUUAAAAAUAAU -----------...(((-.(((((((((((--((((......--...)))))))))...)))).......((((....))))((((((........)))))))))))............. ( -24.80, z-score = -1.01, R) >droSim1.chr3R 24858639 104 - 27517382 -----------GCGGCU-CGCGUCGAAUUG--UUAUGGCCAA--CGAAUGACAGUUAAGUGACAUUUAACGGUUAAUAAGCCUGACAGAUUUACAACUGUCAGCGGCGUUUAAAAAUAAU -----------((.(((-(((....(((((--((((......--...)))))))))..))))........((((....))))((((((........)))))))).))............. ( -26.40, z-score = -1.23, R) >droSec1.super_4 4043862 104 - 6179234 -----------GCGGCU-CGCGUCGAAUUG--UUAUGGCCAA--CGAAUGACAGUUAAGUGACAUUUAACGGUUAAUAAGCCUGACAGAUUUACAACUGUCAGCGACGUUUAAAAAUAAU -----------(((..(-((((((((((((--((((......--...)))))))))...)))).......((((....))))((((((........)))))))))))))........... ( -25.30, z-score = -1.26, R) >droYak2.chr3R 7529013 111 + 28832112 ----GCGGCUCUCGGCU-CGCGUCGAAUUG--UUAUGGCCAA--CGAAUGACAGUUAAGUGACGUUUAACGGUUAAUAAGCCUGACAGAUUUACAACUGUCAGCAACGUUUAAAAAUAAU ----((((((.(((...-.(((((((((((--((((......--...)))))))))...))))))....)))......))))((((((........))))))))................ ( -27.70, z-score = -0.87, R) >droEre2.scaffold_4820 7630230 104 + 10470090 -----------ACGGCU-CGCGUUGAAUUG--UUAUGGCCAG--CGAAUGACAGUUAAGUGACGUUUAACGGUUAAUAAGCCUGACAGAUUUACAACUGUCAGCAACGUUUAAAAAUAAU -----------..((((-.(((......))--)...))))((--((..((((.((((((.....)))))).))))....((.((((((........))))))))..)))).......... ( -24.10, z-score = -0.68, R) >droAna3.scaffold_12911 4875884 104 - 5364042 -----------ACGGCU-AUUGUCGUAUUG--UUAUGGCCGA--CGAAUGACAGUUAAGUGACAUUUAACGGUUAAUAAGCCUGACAGAUUUACAACUGUCAGCGACGUUUAAAAAUAAU -----------(((((.-...)))))((((--((.((((((.--.(((((.((......)).)))))..)))))).((((((((((((........)))))))....)))))..)))))) ( -26.60, z-score = -2.05, R) >dp4.chr2 2040355 111 - 30794189 ---UCUUCUUUGUCGCU-GCUGCUACUCUG---UAUAGCCGA--CGAAUGACACUUAAGUGACAUUUAACGGUUAAUAAGUCUGACAGAUUUACAACUGUCAGCGACGUUUAAAAAUAAU ---........((((((-......(((.((---(.((((((.--.(((((.(((....))).)))))..))))))))))))..(((((........)))))))))))............. ( -29.80, z-score = -2.96, R) >droPer1.super_7 2208053 111 - 4445127 ---UCUUCUUUGUCGCU-GCUGCUACUCUG---UAUAGCCGA--CGAAUGACACUUAAGUGACAUUUAACGGUUAAUAAGUCUGACAGAUUUACAACUGUCAGCGACGUUUAAAAAUAAU ---........((((((-......(((.((---(.((((((.--.(((((.(((....))).)))))..))))))))))))..(((((........)))))))))))............. ( -29.80, z-score = -2.96, R) >droWil1.scaffold_181089 7956516 119 - 12369635 UUGUUUCUGUUGUUGCU-GUUGUUCCUGCGGACUGUGACCAACUCAGAUGACAGUUAAGUGACAUUUAACGGUUAAUAAGCCUGACAGAUUUACAACUGUCAGCGGCUUUUAAAAAUAAC ........(((((((((-(((((..((...((((((.(.(......).).)))))).))..)))....)))))....(((((((((((........))))))..))))).....)))))) ( -29.00, z-score = -0.90, R) >droVir3.scaffold_12822 2513145 109 - 4096053 ----CUUUGUUGUUGUUGGCUGUUGUUGU------UGACCAA-GCGCAUGACAGUUAAGUGACAUUUAACGGUUAAUAAGCCUGACAGAUUUACAACUGUCAGCGACGUUUAAAAAUAAU ----.((((.((((((((((.(((((.((------(...((.-((...((((.(((((((...))))))).))))....)).))...)))..))))).))))))))))..))))...... ( -28.70, z-score = -1.54, R) >consensus ___________GCGGCU_CGCGUCGAAUUG__UUAUGGCCAA__CGAAUGACAGUUAAGUGACAUUUAACGGUUAAUAAGCCUGACAGAUUUACAACUGUCAGCGACGUUUAAAAAUAAU ....................((((...........(((((.....(((((.((......)).)))))...)))))....(.(((((((........))))))))))))............ (-17.13 = -16.51 + -0.62)
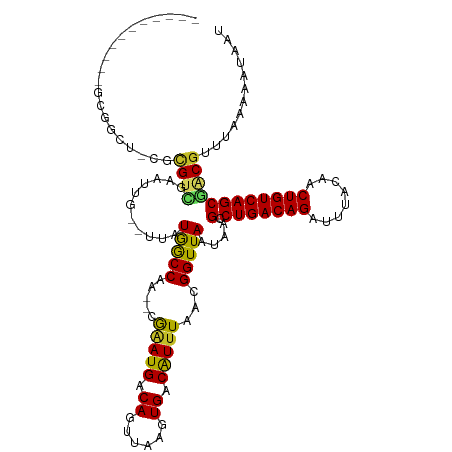
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:11 2011