
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,179,579 – 25,179,672 |
| Length | 93 |
| Max. P | 0.533310 |

| Location | 25,179,579 – 25,179,672 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 63.27 |
| Shannon entropy | 0.73123 |
| G+C content | 0.41082 |
| Mean single sequence MFE | -21.33 |
| Consensus MFE | -6.10 |
| Energy contribution | -6.59 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.533310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
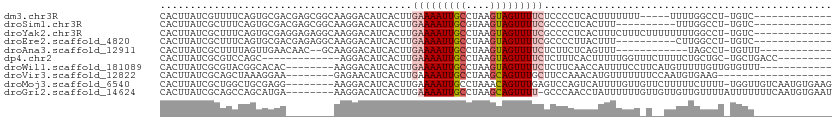
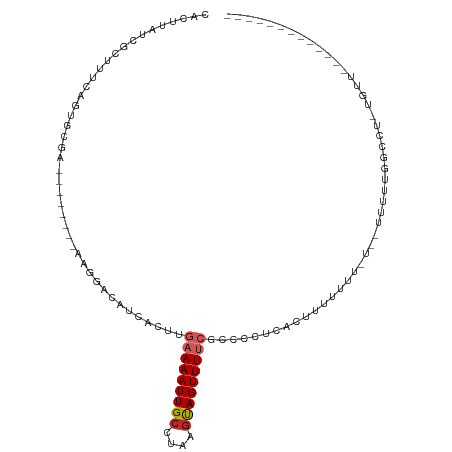
>dm3.chr3R 25179579 93 + 27905053 CACUUAUCGUUUUCAGUGCGACGAGCGGCAAGGACAUCACUUGAAAAUUGCCUAAGUAGUUUUCUCCCCUCACUUUUUUU-----UUUUGGCCU-UGUC------------- ..((..((((.......))))..)).(((((((.........(((((((((....)))))))))...((...........-----....)))))-))))------------- ( -21.16, z-score = -1.52, R) >droSim1.chr3R 24854508 88 + 27517382 CACUUAUCGCUUUCAGUGCGACGAGCGGCAAGGACAUCACUUGAAAAUUGCGUAAGUAGUUUUCGCCCCUCACUUU----------UUUGGCCU-UGUC------------- ..((..((((.......))))..)).(((((((.........(((((((((....)))))))))...((.......----------...)))))-))))------------- ( -27.40, z-score = -2.75, R) >droYak2.chr3R 7524706 98 - 28832112 CACUUAUCGCUUUCAGUGCGAGGAGAGGCAAGGACAUCACUUGAAAAUUGCCUAAGUAGUUUUCGCCCCUCACUUUCUUUCUUUUUUUUGGCCU-UGUC------------- ..(((.((((.......)))).))).(((((((.........(((((((((....)))))))))...((....................)))))-))))------------- ( -23.35, z-score = -0.87, R) >droEre2.scaffold_4820 7626092 88 - 10470090 CACUUAUCGCUUUCAGUGCGACGAGAGGCAAGGACAUCACUUGAAAAUUGCCUAAGUAGUUUUCGCCCCUUACUUU----------CUUGGCCU-UGUC------------- ..((..((((.......))))..)).(((((((.........(((((((((....)))))))))...((.......----------...)))))-))))------------- ( -24.80, z-score = -2.15, R) >droAna3.scaffold_12911 4871776 85 + 5364042 CACUUAUCGCUUUUAGUUGAACAAC--GCAAGGACAUCACUUGAAAAUUGCCUAAGUAGUUUUCUCUUCUCAGUUU------------UAGCCU-UGUUU------------ ..................((((((.--((.(((((.......(((((((((....)))))))))........))))------------).)).)-)))))------------ ( -16.16, z-score = -1.16, R) >dp4.chr2 2036004 89 + 30794189 CACUUAUCGCGUCCAGC-------------AGGACAUCACUUGAAAAUUGCCUAAGUAGUUUUCUCUUUCACUUUUUGGUUUCUUUUCUGCUGC-UGCUGACC--------- ........(((..((((-------------((((........(((((((((....)))))))))......(((....))).....)))))))).-))).....--------- ( -20.40, z-score = -1.93, R) >droWil1.scaffold_181089 7950922 92 + 12369635 CACUUAUCGCGUACGGCACAC--------AAGGACAUCACUUGAAAAUUGCCUAAGUAGUUUUCUCUUCAACCAUUUUCCUUCAUGUUUUUGUUGUGUUU------------ ..............(((((((--------((((((((.....(((((((((....)))))))))...................))))))))).)))))).------------ ( -18.05, z-score = -1.75, R) >droVir3.scaffold_12822 2500720 85 + 4096053 CACUUAUCGCAGCUAAAGGAA--------GAGAACAUCACUUGAAAAUUGCCUAAGCAGUUUGCUUCCAAACAUGUUUUUUUCCAAUGUGAAG------------------- ......(((((......((((--------((((((((...(((.(((((((....))))))).....)))..))))))))))))..)))))..------------------- ( -22.20, z-score = -2.80, R) >droMoj3.scaffold_6540 26610037 103 + 34148556 CACUUAUCGCUGGCUGCGAGG--------AAGGACAUCACUUGAAAAUUGCCUAAACAGUUUGAGUCCAGUCAUUUUGUUGUUCUUUUUCUUUU-UGGUUGUCAAUGUGAAG ..(((.((((.....))))..--------)))....((((((((.....(((.(((.((...(((..(((.(.....))))..)))...)).))-))))..)))).)))).. ( -21.90, z-score = 0.26, R) >droGri2.scaffold_14624 3415010 103 + 4233967 CACUUAUCGCAGCCAGCAUGA--------AAGGACAUCACUUGAAAAUUGCCUAAGCAGUUUU-GCCCAACCUAUUUUUUGUUGUUGUUGUUUUAUUUUUUUCAAUGUGAAU ......(((((......((((--------((.((((.......((((((((....))))))))-...((((.........)))).)))).)))))).........))))).. ( -17.86, z-score = 0.05, R) >consensus CACUUAUCGCUUUCAGUGCGA________AAGGACAUCACUUGAAAAUUGCCUAAGUAGUUUUCGCCCCUCACUUUUUUU_U__UUUUUGGCCU_UGUU_____________ ..........................................(((((((((....)))))))))................................................ ( -6.10 = -6.59 + 0.49)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:08 2011