
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,169,893 – 25,169,983 |
| Length | 90 |
| Max. P | 0.985861 |

| Location | 25,169,893 – 25,169,983 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.39 |
| Shannon entropy | 0.47292 |
| G+C content | 0.47290 |
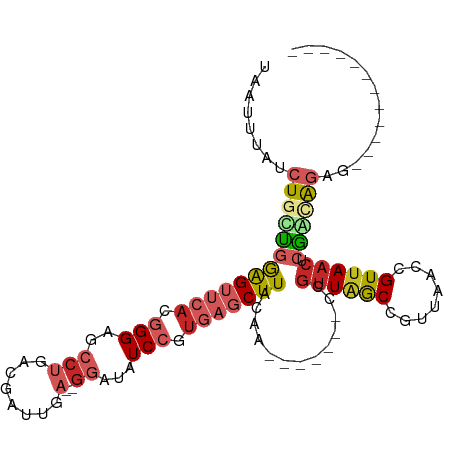
| Mean single sequence MFE | -30.24 |
| Consensus MFE | -14.05 |
| Energy contribution | -15.47 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.985861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
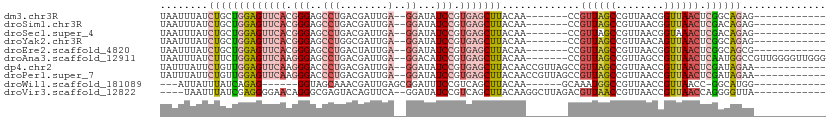
>dm3.chr3R 25169893 90 - 27905053 UAAUUUAUCUGCUGGAGUUCACGGGAGCCUGACGAUUGA--GGAUAUCCGUGAGCUUACAA-------CCGUUAGCCGUUAACGGUUAACUCGGCAGAG------------ .......((((((((((((((((((..(((........)--))...)))))))))))....-------..((((((((....)))))))).))))))).------------ ( -41.50, z-score = -6.25, R) >droSim1.chr3R 24844862 90 - 27517382 UAAUUUAUCUGCUGGAGUUCACGGGAGCCUGACGAUUGA--GGAUAUCCGUGAGCUUACAA-------CCGUUAGCCGUUAACGGUUAACUCGACAGAG------------ .......((((.(((((((((((((..(((........)--))...)))))))))))....-------..((((((((....)))))))).)).)))).------------ ( -34.90, z-score = -4.58, R) >droSec1.super_4 4030220 90 - 6179234 UAAUUUAUCUGCUGGAGUUCACGGGAGCCUGACGAUUGA--GGAUAUCCGUGAGCUUACAA-------CCGUUAGCCGUUAACGGUAAACUCGACAGAG------------ .......((((.(((((((((((((..(((........)--))...)))))))))))....-------..(((.((((....)))).))).)).)))).------------ ( -31.30, z-score = -3.43, R) >droYak2.chr3R 7513813 90 + 28832112 UAAUUUAUCUGCUGGAGUUCACGGGAGCCUGGCGAUUGA--GGAUAUCCGUGAGCUUACAA-------CCGUUAGCCGUUAACAGUUAACUCGGCAGAG------------ .......((((((((((((((((((..(((........)--))...)))))))))))....-------..((((((.(....).)))))).))))))).------------ ( -35.40, z-score = -4.17, R) >droEre2.scaffold_4820 7616597 90 + 10470090 UAAUUUAUCUGCUGGAGUUCACGGGAGCCUGACUAUUGA--GGAUAUCCGUGAGCUUACAA-------CCGUUAGCCGUUAACGGUUAACUCGGCAGCG------------ ........(((((((((((((((((..(((........)--))...)))))))))))....-------..((((((((....)))))))).))))))..------------ ( -39.00, z-score = -5.35, R) >droAna3.scaffold_12911 4861724 102 - 5364042 UAAUUUAUCUUCUGGAGUUCAAGGGAGCCUGACGAUUGA--GGACAUCCGUGAGCUUACAA-------CCGUUAGCCGUUAGCCGUUAACUCAAUGGCCGUUGGGGUUGGG ..............(((((((..(((.(((........)--))...))).))))))).(((-------((((((((.(....).))))))(((((....)))))))))).. ( -29.50, z-score = -0.28, R) >dp4.chr2 2022755 97 - 30794189 UAUUUAUUCUGUUGGAGUUCAAGGGACCCUGACGAUUGA--GGAUAUCCGUGAGCUUACAACCGUUAGCCGUUAGCCGUUAACCGUUAACUCGAUAGAA------------ ......(((((((((((((((.(((..(((........)--))...))).)))))))......((((((.(((((...))))).)))))).))))))))------------ ( -27.30, z-score = -1.98, R) >droPer1.super_7 2190389 97 - 4445127 UAUUUAUUCUGUUGGAGUUCAAGGGACCCUGACGAUUGA--GGAUAUCCGUGAGCUUACAACCGUUAGCCGUUAGCCGUUAACCGUUAACUCGAUAGAA------------ ......(((((((((((((((.(((..(((........)--))...))).)))))))......((((((.(((((...))))).)))))).))))))))------------ ( -27.30, z-score = -1.98, R) >droWil1.scaffold_181089 7939108 83 - 12369635 ---AUUAUUUAUCAGAG------GGUAGCAAACGAUUGAGCGGAUUUCCGUCAGCUUACAA------GCAAAUGGCCGUUAACCGUUAACC-GGCAUGG------------ ---...........(..------(((....((((....(((((....((((..(((....)------))..)))))))))...)))).)))-..)....------------ ( -16.00, z-score = 0.65, R) >droVir3.scaffold_12822 2485427 93 - 4096053 ----UAAUUUAUCGAGGGGAACAGGGCGAGUACAGUUCA--GGAUAUCCGUCAGCUUACAAGGCUUAGACGUUAACCGUUAACCGUUAACCAGGGGUUA------------ ----...........((..(((.((((.......)))).--((.((..((((((((.....))))..)))).)).)))))..))..(((((...)))))------------ ( -20.20, z-score = 0.06, R) >consensus UAAUUUAUCUGCUGGAGUUCACGGGAGCCUGACGAUUGA__GGAUAUCCGUGAGCUUACAA_______CCGUUAGCCGUUAACCGUUAACUCGACAGAG____________ ........(((((((((((((.(((..((............))...))).))))))).............((((((........)))))).)))))).............. (-14.05 = -15.47 + 1.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:05 2011