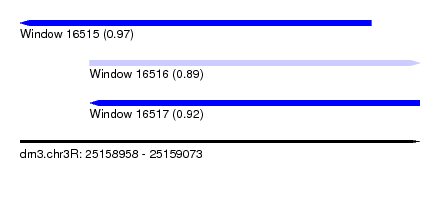
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,158,958 – 25,159,073 |
| Length | 115 |
| Max. P | 0.974585 |

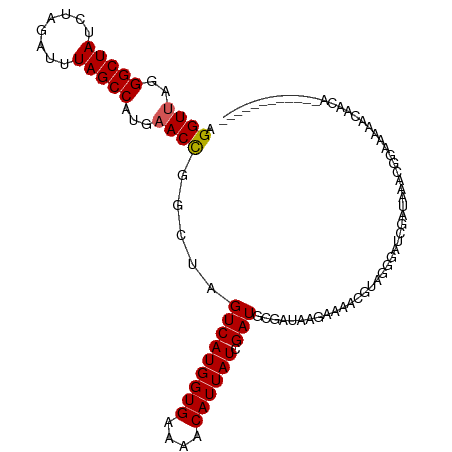
| Location | 25,158,958 – 25,159,059 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 70.93 |
| Shannon entropy | 0.51172 |
| G+C content | 0.41419 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -16.92 |
| Energy contribution | -16.88 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.974585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25158958 101 - 27905053 AGGUUAGGGCUAUCUAGAUUUAGCCAUGAACCGGCUAGUCAUGGUGAAAACAUUAUCGAUGCGAUAAGAAAACCUAGGGAUUCCUGAUCUUAAAAACAUCA------------ (((((((((...(((((.(((.(((((((.(......)))))))).)))...((((((...))))))......)))))...)))))))))...........------------ ( -26.50, z-score = -1.79, R) >droEre2.scaffold_4820 7606027 113 + 10470090 AGGUUGGGGCUAUCUGGAUUUAGCCAUGAACUGGCUAGUCAUGGUGCAAACAUUAUUGAUACCAUGGGAAAAUGUAGGGCUCGAAAAAUGGUUUCCUCACCUUCAAAACAUCA ((((.((((..(((.....(((((((.....)))))))(((((((((((......))).))))))))......................)))..))))))))........... ( -29.00, z-score = -0.30, R) >droYak2.chr3R 7502531 112 + 28832112 AGGUGAGGGCUAUCUAGAUUUAGCCAUGAACUGGCUAGUCAUGGUGAAAACAUUAUUGAUACCAUAAGAAAAUGCGGAA-UCGACAAAUGGAUUCCUGACCUAAAAAACAUCA ((((.((((..(((((....((((((.....))))))(((((((((....)))))).(((.((............)).)-)))))...))))))))).))))........... ( -28.90, z-score = -2.33, R) >droSec1.super_4 4019553 101 - 6179234 AGGUUGGGGCUAUCUAGAUUUAGCCAUGAACCGGCUAGUCAUGGUGGAAACAUUAUCGAUGCGAUAAGAAAACGUAGCGAUUCCUGACCUUAAAAACAUCA------------ ((((..(((.......((((.((((.......)))))))).(((((....)))))(((.((((.........)))).))).)))..))))...........------------ ( -28.80, z-score = -2.15, R) >droSim1.chrU 1044333 84 + 15797150 AGGUUAGGGCUAUCUAGAUUUAGCCGUGAACCGGCUAGUCAUGGUGGAAACAUUAUCGAUGCGAUAAGAAAACGUAGGGAUAGA----------------------------- .........((((((.((((.(((((.....)))))))))((((((....))))))...((((.........)))).)))))).----------------------------- ( -23.30, z-score = -2.05, R) >consensus AGGUUAGGGCUAUCUAGAUUUAGCCAUGAACCGGCUAGUCAUGGUGAAAACAUUAUCGAUGCGAUAAGAAAACGUAGGGAUCGAUAAACGGAAAAACAACA____________ .((((..(((((........)))))...)))).....(((((((((....)))))).)))..................................................... (-16.92 = -16.88 + -0.04)

| Location | 25,158,978 – 25,159,073 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.77 |
| Shannon entropy | 0.31333 |
| G+C content | 0.44851 |
| Mean single sequence MFE | -24.69 |
| Consensus MFE | -18.50 |
| Energy contribution | -17.70 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.892796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25158978 95 + 27905053 ------------AUCCCUAGGUUUUCUUAUCGCAUCGAUAAUGUUUUCACCAUGACUAGCCGGUUCAUGGCUAAAUCUAGAUAGCCCUAACCUAGGGCAUAUGGCUC ------------..(((((((((...(((((((((.....))))............((((((.....))))))......)))))....))))))))).......... ( -25.70, z-score = -1.71, R) >droEre2.scaffold_4820 7606047 107 - 10470090 AACCAUUUUUCGAGCCCUACAUUUUCCCAUGGUAUCAAUAAUGUUUGCACCAUGACUAGCCAGUUCAUGGCUAAAUCCAGAUAGCCCCAACCUAGGGCAUACGGCUC ...........(((((...........((((((..(((......))).))))))..((((((.....))))))..........((((.......))))....))))) ( -30.60, z-score = -3.39, R) >droYak2.chr3R 7502551 106 - 28832112 -AUCCAUUUGUCGAUUCCGCAUUUUCUUAUGGUAUCAAUAAUGUUUUCACCAUGACUAGCCAGUUCAUGGCUAAAUCUAGAUAGCCCUCACCUAGGGCAUAUGACUC -........(((((((..........(((((((...............))))))).((((((.....))))))))))......(((((.....)))))....))).. ( -25.06, z-score = -2.06, R) >droSec1.super_4 4019573 95 + 6179234 ------------AUCGCUACGUUUUCUUAUCGCAUCGAUAAUGUUUCCACCAUGACUAGCCGGUUCAUGGCUAAAUCUAGAUAGCCCCAACCUAGGGCAUACGGCUC ------------...(((.........((((((((.....))))............((((((.....))))))......))))((((.......))))....))).. ( -19.90, z-score = -0.18, R) >droSim1.chrU 1044336 95 - 15797150 ------------AUCCCUACGUUUUCUUAUCGCAUCGAUAAUGUUUCCACCAUGACUAGCCGGUUCACGGCUAAAUCUAGAUAGCCCUAACCUAGGGCAUACAGCUC ------------...................(((((......(((........)))((((((.....))))))......))).((((((...)))))).....)).. ( -22.20, z-score = -1.87, R) >consensus ____________AUCCCUACGUUUUCUUAUCGCAUCGAUAAUGUUUCCACCAUGACUAGCCGGUUCAUGGCUAAAUCUAGAUAGCCCUAACCUAGGGCAUACGGCUC ...............................(((((......(((........)))((((((.....))))))......))).((((.......)))).....)).. (-18.50 = -17.70 + -0.80)

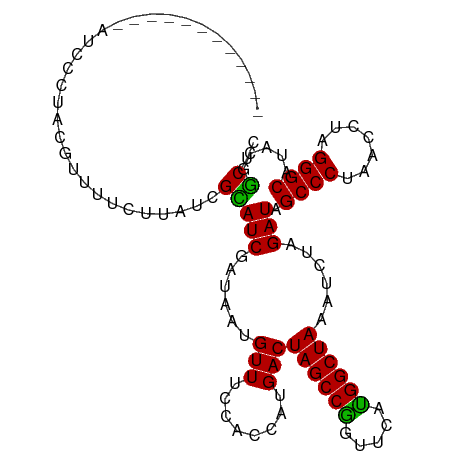
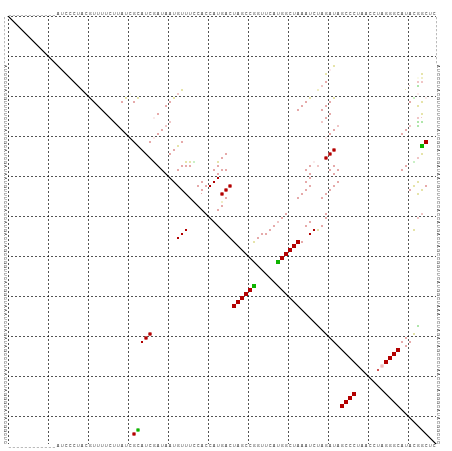
| Location | 25,158,978 – 25,159,073 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.77 |
| Shannon entropy | 0.31333 |
| G+C content | 0.44851 |
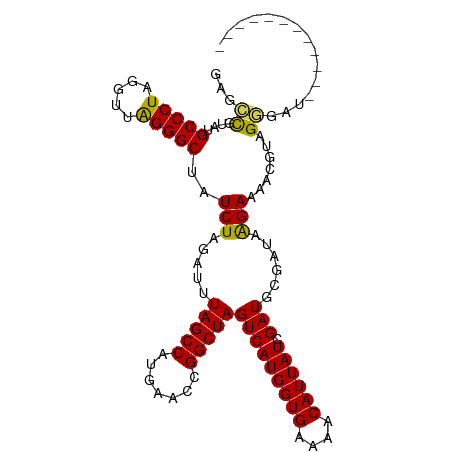
| Mean single sequence MFE | -30.44 |
| Consensus MFE | -22.28 |
| Energy contribution | -21.68 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.921674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25158978 95 - 27905053 GAGCCAUAUGCCCUAGGUUAGGGCUAUCUAGAUUUAGCCAUGAACCGGCUAGUCAUGGUGAAAACAUUAUCGAUGCGAUAAGAAAACCUAGGGAU------------ ..........(((((((((...(((((...((((.((((.......))))))))))))).......((((((...))))))...)))))))))..------------ ( -28.60, z-score = -2.15, R) >droEre2.scaffold_4820 7606047 107 + 10470090 GAGCCGUAUGCCCUAGGUUGGGGCUAUCUGGAUUUAGCCAUGAACUGGCUAGUCAUGGUGCAAACAUUAUUGAUACCAUGGGAAAAUGUAGGGCUCGAAAAAUGGUU (((((.((((((((.....)))))..(((....(((((((.....))))))).((((((((((......))).))))))))))....))).)))))........... ( -39.30, z-score = -3.18, R) >droYak2.chr3R 7502551 106 + 28832112 GAGUCAUAUGCCCUAGGUGAGGGCUAUCUAGAUUUAGCCAUGAACUGGCUAGUCAUGGUGAAAACAUUAUUGAUACCAUAAGAAAAUGCGGAAUCGACAAAUGGAU- ..(((....(((((.....)))))..(((.((((.(((((.....)))))))))((((((..((.....))..)))))).)))............)))........- ( -26.50, z-score = -1.34, R) >droSec1.super_4 4019573 95 - 6179234 GAGCCGUAUGCCCUAGGUUGGGGCUAUCUAGAUUUAGCCAUGAACCGGCUAGUCAUGGUGGAAACAUUAUCGAUGCGAUAAGAAAACGUAGCGAU------------ ..(((((..(((((.....)))))((((......(((((.......)))))(((((((((....)))))).)))..)))).....)))..))...------------ ( -28.00, z-score = -1.33, R) >droSim1.chrU 1044336 95 + 15797150 GAGCUGUAUGCCCUAGGUUAGGGCUAUCUAGAUUUAGCCGUGAACCGGCUAGUCAUGGUGGAAACAUUAUCGAUGCGAUAAGAAAACGUAGGGAU------------ ...((.(((((((((...))))))((((......((((((.....))))))(((((((((....)))))).)))..)))).......))).))..------------ ( -29.80, z-score = -2.05, R) >consensus GAGCCGUAUGCCCUAGGUUAGGGCUAUCUAGAUUUAGCCAUGAACCGGCUAGUCAUGGUGAAAACAUUAUCGAUGCGAUAAGAAAACGUAGGGAU____________ ...((....(((((.....)))))..(((.....(((((.......)))))(((((((((....)))))).)))......))).......))............... (-22.28 = -21.68 + -0.60)

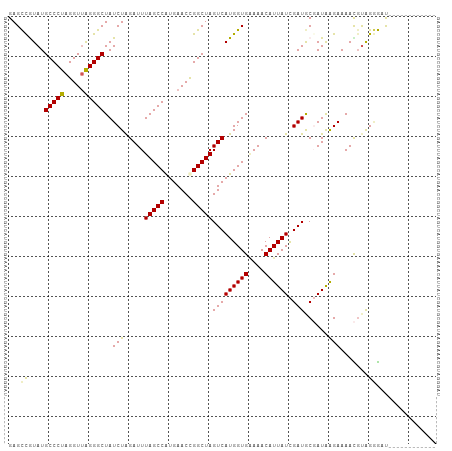
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:05 2011