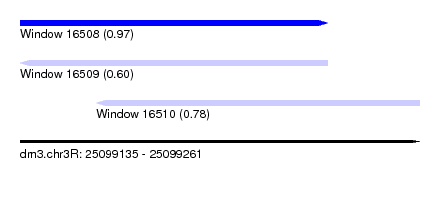
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,099,135 – 25,099,261 |
| Length | 126 |
| Max. P | 0.971502 |

| Location | 25,099,135 – 25,099,232 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 78.65 |
| Shannon entropy | 0.40504 |
| G+C content | 0.50299 |
| Mean single sequence MFE | -25.01 |
| Consensus MFE | -16.97 |
| Energy contribution | -16.79 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
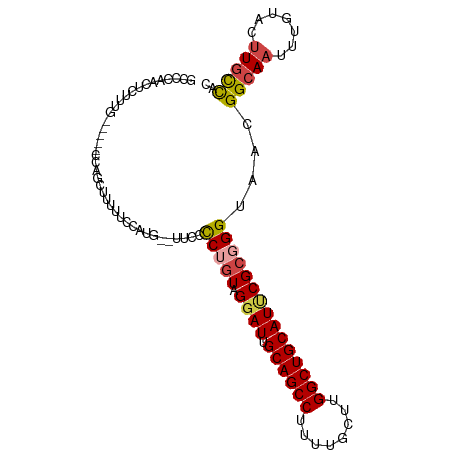
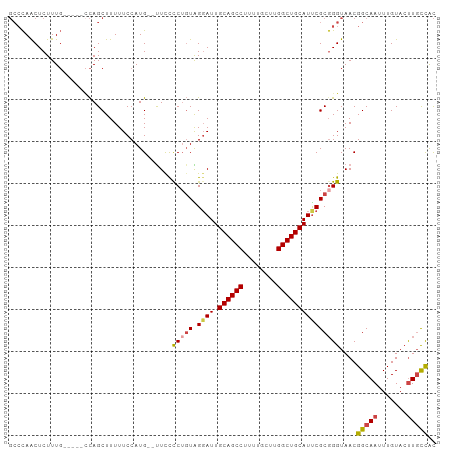
>dm3.chr3R 25099135 97 + 27905053 ACCCAAAAGCAGAAGGCUAAACCCGUGGCAAGUACAAAUUGCCGUUACCGGCGAAUGCAGCCAAGCAAAAGGCUGCAAUCCUACAGGGGAAAACAUG .(((...(((.....)))......((((..........((((((....)))))).(((((((........)))))))...)))).)))......... ( -31.60, z-score = -2.34, R) >droSim1.chr3R 24772967 97 + 27517382 ACCCAAAAGCAGAAGGCUAAACCCGUGGCAAGUACAAAUUGCCGUUACCCGCGAAUGCAGCCAAGCAAAAGGCUGCAAUCCUACAGGGGAAACCAUG .(((....((....((......))..(((((.......))))).......))((.(((((((........))))))).)).....)))......... ( -29.60, z-score = -1.93, R) >droSec1.super_4 3959940 97 + 6179234 ACCCAAAAGCAGAAGGCUAAACCCGUGGCAAGUACAAAUUGCCGUUACCCGCGAAUGCAGCCAAGCAAAAGGCUGCAAUACUACAGGGGAAGCCAUG ..............((((...(((..(((((.......)))))............(((((((........)))))))........)))..))))... ( -32.20, z-score = -2.43, R) >droEre2.scaffold_4820 7543448 95 - 10470090 ACCCAAAAGCAGAGAGCGAAACCCGUGGCAAGUACAAAUUGCCGUUACCCGCGGAUGCAGCCAAGCAAAAGGCUGCAAUCCCACUGGGAAG--CAUG .((((...((.....(((.....)))(((((.......))))).......))((((((((((........)))))).))))...))))...--.... ( -33.10, z-score = -2.91, R) >droAna3.scaffold_12911 4788047 96 + 5364042 UCCUAAAGGUAGAAACCCAAAGCCAAAACAACUACAAAUUGCCGUUACCCGCGAAUGCAACCAAGCAAAAGGCUGCAAUCCU-CAAGGAAAACAUUG ((((..(((...........((((..............((((........)))).(((......)))...)))).....)))-..))))........ ( -16.59, z-score = -1.02, R) >dp4.chr2 1938108 90 + 30794189 GCAUAAACCCAAGUCCCAAAUCCCACAAGAGCUACAAAUUGCCGUUACCCGCGAAUGCAGCCAAGC----GGCUGCAAUCCC---AAGAAAAACAUG ((..................((......))((........))........))((.(((((((....----))))))).))..---............ ( -16.00, z-score = -1.89, R) >droPer1.super_7 2102199 90 + 4445127 GCAUAAACCCAAGUCCCAAAUCCCACAAGAGCUACAAAUUGCCGUUACCCGCGAAUGCAGCCAAGC----GGCUGCAAUCCC---AAGAAAAACAUG ((..................((......))((........))........))((.(((((((....----))))))).))..---............ ( -16.00, z-score = -1.89, R) >consensus ACCCAAAAGCAGAAGGCUAAACCCGUGGCAAGUACAAAUUGCCGUUACCCGCGAAUGCAGCCAAGCAAAAGGCUGCAAUCCUACAGGGAAAAACAUG .(((..................................((((........)))).(((((((........)))))))........)))......... (-16.97 = -16.79 + -0.18)
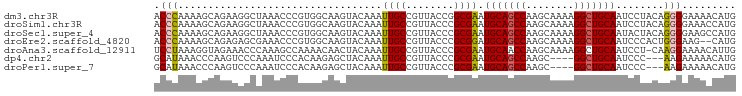
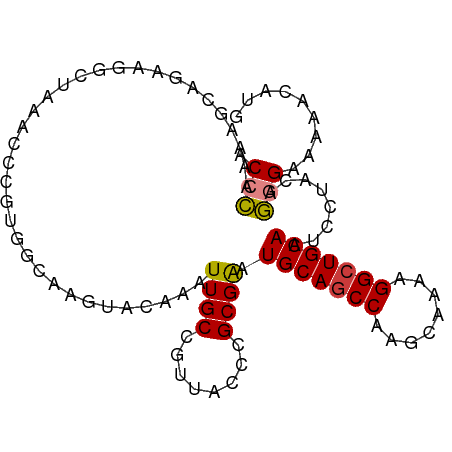
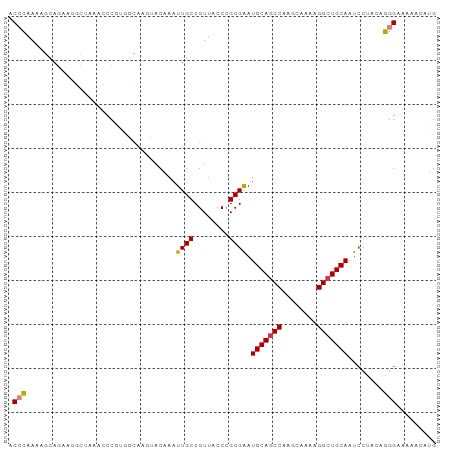
| Location | 25,099,135 – 25,099,232 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 78.65 |
| Shannon entropy | 0.40504 |
| G+C content | 0.50299 |
| Mean single sequence MFE | -30.99 |
| Consensus MFE | -19.38 |
| Energy contribution | -18.81 |
| Covariance contribution | -0.57 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.602807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25099135 97 - 27905053 CAUGUUUUCCCCUGUAGGAUUGCAGCCUUUUGCUUGGCUGCAUUCGCCGGUAACGGCAAUUUGUACUUGCCACGGGUUUAGCCUUCUGCUUUUGGGU .........(((.((((((.(((((((........))))))).))(((.((...(((((.......))))))).)))........))))....))). ( -33.40, z-score = -1.79, R) >droSim1.chr3R 24772967 97 - 27517382 CAUGGUUUCCCCUGUAGGAUUGCAGCCUUUUGCUUGGCUGCAUUCGCGGGUAACGGCAAUUUGUACUUGCCACGGGUUUAGCCUUCUGCUUUUGGGU .........(((.....((.(((((((........))))))).))(((((....(((((.......)))))..(((.....))))))))....))). ( -33.40, z-score = -1.16, R) >droSec1.super_4 3959940 97 - 6179234 CAUGGCUUCCCCUGUAGUAUUGCAGCCUUUUGCUUGGCUGCAUUCGCGGGUAACGGCAAUUUGUACUUGCCACGGGUUUAGCCUUCUGCUUUUGGGU ...((((.((((((..(...(((((((........))))))).)..))).....(((((.......)))))..)))...)))).............. ( -33.90, z-score = -1.29, R) >droEre2.scaffold_4820 7543448 95 + 10470090 CAUG--CUUCCCAGUGGGAUUGCAGCCUUUUGCUUGGCUGCAUCCGCGGGUAACGGCAAUUUGUACUUGCCACGGGUUUCGCUCUCUGCUUUUGGGU ....--...(((((..((((.((((((........))))))))))(((((....(((((.......)))))..(((.....))))))))..))))). ( -36.50, z-score = -2.02, R) >droAna3.scaffold_12911 4788047 96 - 5364042 CAAUGUUUUCCUUG-AGGAUUGCAGCCUUUUGCUUGGUUGCAUUCGCGGGUAACGGCAAUUUGUAGUUGUUUUGGCUUUGGGUUUCUACCUUUAGGA ........((((.(-(((..(((((((........)))))))...((.((.((((((........)))))))).))............)))).)))) ( -27.50, z-score = -1.36, R) >dp4.chr2 1938108 90 - 30794189 CAUGUUUUUCUU---GGGAUUGCAGCC----GCUUGGCUGCAUUCGCGGGUAACGGCAAUUUGUAGCUCUUGUGGGAUUUGGGACUUGGGUUUAUGC ..(((..(((((---(.((.(((((((----....))))))).)).)))).))..)))......(((((..((..........))..)))))..... ( -26.10, z-score = -0.92, R) >droPer1.super_7 2102199 90 - 4445127 CAUGUUUUUCUU---GGGAUUGCAGCC----GCUUGGCUGCAUUCGCGGGUAACGGCAAUUUGUAGCUCUUGUGGGAUUUGGGACUUGGGUUUAUGC ..(((..(((((---(.((.(((((((----....))))))).)).)))).))..)))......(((((..((..........))..)))))..... ( -26.10, z-score = -0.92, R) >consensus CAUGGUUUUCCCUGUAGGAUUGCAGCCUUUUGCUUGGCUGCAUUCGCGGGUAACGGCAAUUUGUACUUGCCACGGGUUUAGCCUUCUGCUUUUGGGU .........(((....((((.((((((........))))))))))((.(....).))...........((...(((.....)))...))....))). (-19.38 = -18.81 + -0.57)
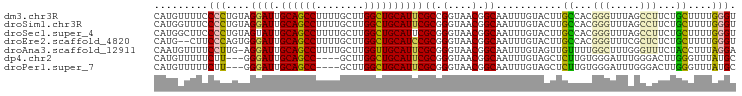
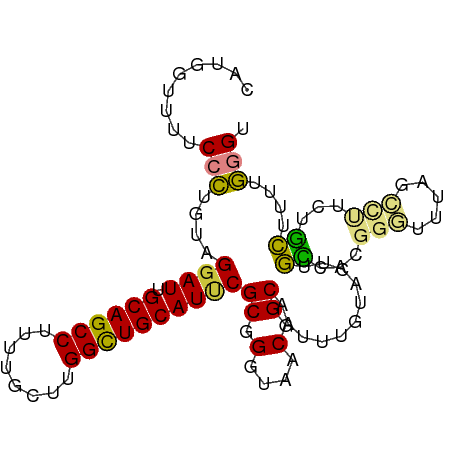
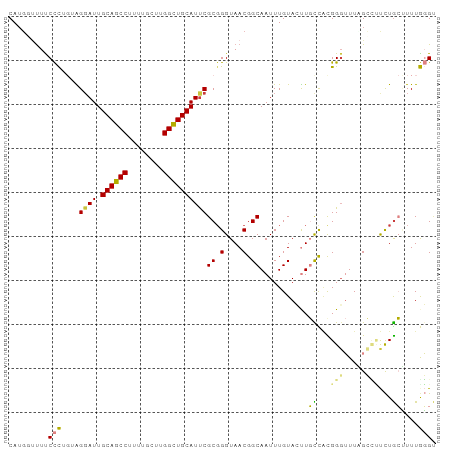
| Location | 25,099,159 – 25,099,261 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 85.02 |
| Shannon entropy | 0.27970 |
| G+C content | 0.50937 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -21.96 |
| Energy contribution | -22.43 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.781759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
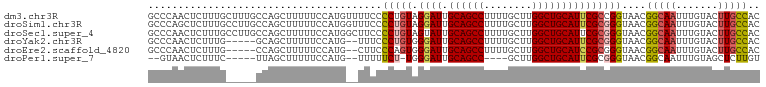
>dm3.chr3R 25099159 102 - 27905053 GCCCAACUCUUUGCUUUGCCAGCUUUUUCCAUGUUUUCCCCUGUAGGAUUGCAGCCUUUUGCUUGGCUGCAUUCGCCGGUAACGGCAAUUUGUACUUGCCAC ((...((...((((((((((.((.............(((......))).(((((((........)))))))...)).))))).)))))...))....))... ( -29.30, z-score = -1.90, R) >droSim1.chr3R 24772991 102 - 27517382 GCCCAGCUCUUUGCCUUGCCAGCUUUUUCCAUGGUUUCCCCUGUAGGAUUGCAGCCUUUUGCUUGGCUGCAUUCGCGGGUAACGGCAAUUUGUACUUGCCAC ((...((...((((((((((.((......((.((....)).))...((.(((((((........))))))).)))).))))).)))))...))....))... ( -33.90, z-score = -1.57, R) >droSec1.super_4 3959964 102 - 6179234 GCCCAACUCUUUGCCUUGCCAGCUUUUUCCAUGGCUUCCCCUGUAGUAUUGCAGCCUUUUGCUUGGCUGCAUUCGCGGGUAACGGCAAUUUGUACUUGCCAC ((...((...(((((..((((..........))))....((((..(...(((((((........))))))).)..))))....)))))...))....))... ( -32.20, z-score = -1.89, R) >droYak2.chr3R 7440748 95 + 28832112 GCCCAACUCUUUG-----GCAGCUUUUUCCAUG--UUUCCCUGUGGGAUUGCAGCCUUUUGCUUGGCUGCAUUCGCGGGUAACGGCAAUUUGUACUUGCCAC (((.........)-----))............(--((..((((((((..(((((((........))))))))))))))).)))(((((.......))))).. ( -32.90, z-score = -2.11, R) >droEre2.scaffold_4820 7543472 95 + 10470090 GCCCAACUCUUUG-----CCAGCUUUUUCCAUG--CUUCCCAGUGGGAUUGCAGCCUUUUGCUUGGCUGCAUCCGCGGGUAACGGCAAUUUGUACUUGCCAC ((...((...(((-----(((((.........)--)).(((.(((((..(((((((........)))))))))))))))....)))))...))....))... ( -33.40, z-score = -2.13, R) >droPer1.super_7 2102223 88 - 4445127 --GUAACUCUUUC-----UUAGCUUUUUCCAUG--UUUUUCU-UGGGAUUGCAGCC----GCUUGGCUGCAUUCGCGGGUAACGGCAAUUUGUAGCUCUUGU --...........-----..((((.......((--(..((((-((.((.(((((((----....))))))).)).)))).))..)))......))))..... ( -25.62, z-score = -1.97, R) >consensus GCCCAACUCUUUG_____CCAGCUUUUUCCAUG__UUCCCCUGUAGGAUUGCAGCCUUUUGCUUGGCUGCAUUCGCGGGUAACGGCAAUUUGUACUUGCCAC .......................................(((((.((((.((((((........)))))))))))))))....(((((.......))))).. (-21.96 = -22.43 + 0.47)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:59 2011