
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,097,091 – 25,097,197 |
| Length | 106 |
| Max. P | 0.658773 |

| Location | 25,097,091 – 25,097,197 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 88.86 |
| Shannon entropy | 0.20730 |
| G+C content | 0.41692 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -22.42 |
| Energy contribution | -22.62 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.658773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25097091 106 + 27905053 GUGUCAUGGGGUUUUGUUUGUGUUUUUU-CGCAAUUGAUUUGCGGAGAAAGCAUCGAUUGUAGGGCAAUUGUGCGAAAAUUGGGAAAAGUCACAGGAGACAUGAGCA------------ (((((.((((.((((.(..((.((((((-(((((.....)))))))))))(((.((((((.....)))))))))....))..).)))).)).))...))))).....------------ ( -29.50, z-score = -1.92, R) >droSim1.chr3R 24770845 105 + 27517382 GUGUCAUGGGGUUUUGUUUGUGUUUUAU-CGCA-UUGAUUUGCGGAGAAAGCAUCGAUUGUAGGGCAAUUGUGCGAAAAUUGGGAAAAGUCACAGGAGACAUGAGCG------------ ...............(((..((((((.(-((((-......))))).....(((.((((((.....))))))))).....................))))))..))).------------ ( -25.70, z-score = -0.75, R) >droSec1.super_4 3957871 106 + 6179234 GUGUCAUGGGGUUUUGUUUGUGUUUUAU-CGCAAUUGAUUUGCGGAGAAAGCAUCGAUUGUAGGGCAAUUGUGCGAAAAUUGGGAAAAGUCACAGGAGACAUGAGCG------------ ...............(((..((((((.(-(((((.....)))))).....(((.((((((.....))))))))).....................))))))..))).------------ ( -26.40, z-score = -0.98, R) >droEre2.scaffold_4820 7541375 105 - 10470090 GUGUCAUGGGGUUUUGUUUGUGUUUUUUUCGCAAUUGAUUUGCGGAGAAAGCAUCGAUUGUAGGGCAAUUGUGCGAAAAUUGGGAAAAGUCACAGGAGACAUGCG-------------- (((((.((((.((((....(((((((((((((((.....)))))))))))))))(((((...(.((....)).)...)))))..)))).)).))...)))))...-------------- ( -30.70, z-score = -2.37, R) >droAna3.scaffold_12911 4785931 105 + 5364042 GUGUCAUGGGGUUUUGUUUGUGUUUUUUUCGCAAUUGAUUUGCGGAGAAAGCAUCGAUUGUAGGGCAAUUGUGCAAAAAUUGGAAAAAGUCACAGGAGACAUCCG-------------- (((.(......((((((..(((((((((((((((.....)))))))))))))))((((((.....)))))).)))))).((....)).).))).(((....))).-------------- ( -30.20, z-score = -2.20, R) >dp4.chr2 1935819 118 + 30794189 GUGUCAUGGGGUUUUGUUUGUGUUUUUUC-GCAAUUGAUUUGCGGAGAAAGCAUCGAUUGUAGGGUAAUUGUGCAAAAAUUGGGAAAAGUCACAGGAGACAUCCGCAUCCGCACAUUUU ((((.(((((((.((.(((((((((((((-((((.....)))))))))))(((.((((((.....)))))))))................)))))).)).)))).)))..))))..... ( -32.50, z-score = -1.33, R) >droPer1.super_7 2099904 118 + 4445127 GUGUCAUGGGGUUUUGUUUGUGUUUUUUC-GCAAUUGAUUUGCGGAGAAAGCAUCGAUUGUAGGGUAAUUGUGCAAAAAUUGGGAAAAGUCACAGGAGACAUCCGAAUCCGCACAUUUU ((((....((((((....(((((((((((-((((.....)))))))))))(((.((((((.....)))))))))................))))(((....)))))))))))))..... ( -31.80, z-score = -1.31, R) >droWil1.scaffold_181089 7854725 103 + 12369635 GUGUCAUGGGGUUUUGUUUGUGUUUUUUCUGCAAUUGAUUUGCGGAGAAAGCAUCGAUUGUAGCAUAAUUGUGCAAAAAUUGGAAAAAGUCACAGGGGACAUA---------------- (((((.((((.((((....(((((((((((((((.....)))))))))))))))(((((...((((....))))...)))))..)))).)).))...))))).---------------- ( -33.40, z-score = -4.20, R) >consensus GUGUCAUGGGGUUUUGUUUGUGUUUUUU_CGCAAUUGAUUUGCGGAGAAAGCAUCGAUUGUAGGGCAAUUGUGCAAAAAUUGGGAAAAGUCACAGGAGACAUGCGC_____________ (((((.((((.((((....(((((((((.(((((.....))))).)))))))))(((((...(.((....)).)...)))))..)))).)).))...)))))................. (-22.42 = -22.62 + 0.20)

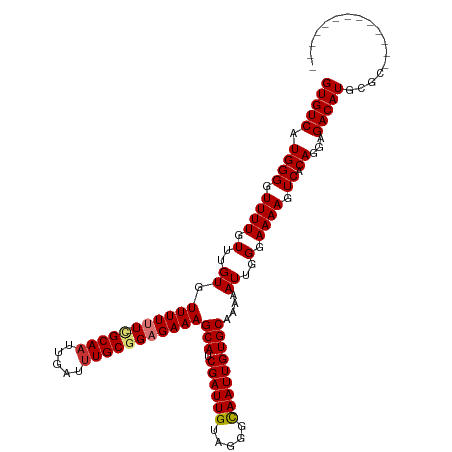

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:56 2011