
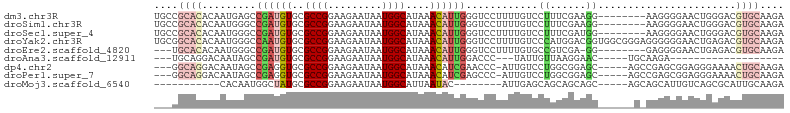
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,092,054 – 25,092,151 |
| Length | 97 |
| Max. P | 0.831858 |

| Location | 25,092,054 – 25,092,151 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 66.23 |
| Shannon entropy | 0.69284 |
| G+C content | 0.52631 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -9.30 |
| Energy contribution | -9.92 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.831858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
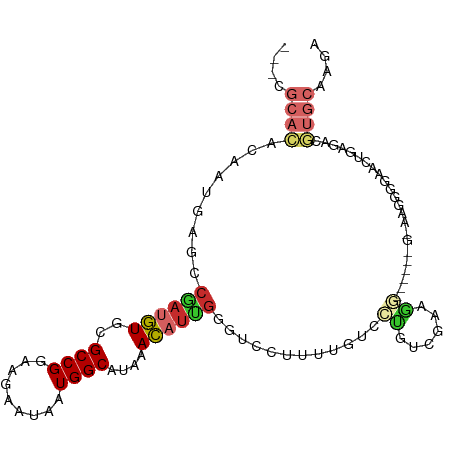
>dm3.chr3R 25092054 97 - 27905053 UGCCGCACACAAUGAGCCGAUGUGCGCCGGAAGAAUAAUGGCAUAAACAUUGGGUCCUUUUGUCCUUUCGAAGG--------AAGGGGAACUGGGACGUGCAAGA ....((((........(((((((..((((.........))))....)))))))(((((....(((((((....)--------))))))....))))))))).... ( -39.10, z-score = -3.75, R) >droSim1.chr3R 24765348 97 - 27517382 UGCCGCACACAAUGGGCCGAUGUGCGCCGGAAGAAUAAUGGCAUAAACAUUGGGUCCUUUUGUCCUUUCGAAGG--------AAGGGGAACUGGGACGUGCAAGA ....((((........(((((((..((((.........))))....)))))))(((((....(((((((....)--------))))))....))))))))).... ( -39.10, z-score = -3.39, R) >droSec1.super_4 3952898 97 - 6179234 UGCCGCACACAAUGGGCCGAUGUGCGCCGGAAGAAUAAUGGCAUAAACAUUGGGUCCUUUUGUCCUUUCGAUGG--------AAGGGGAACUGGGACGUGCAAGA ....((((........(((((((..((((.........))))....)))))))(((((....(((((((....)--------))))))....))))))))).... ( -36.60, z-score = -2.52, R) >droYak2.chr3R 7433447 105 + 28832112 UGCGGCACACAAUGGGCCAAUGUGCGCCGGAAGAAUAAUGGCAUAAACAUUGGGUCCUUUUGUCCCAUGGACGGUGGCGGGAGGGGGGAACUGAGACGUGCAAGA ....((((........(((((((..((((.........))))....))))))).((((((((((((......)).))))))))))............)))).... ( -34.00, z-score = -0.49, R) >droEre2.scaffold_4820 7536391 93 + 10470090 ---UGCACACAAUGGGCCGAUGUGCGCCGGAAGAAUAAUGGCAUAAACAUUGGGUCCUUUUGUGCCGUCGA-GG--------GAGGGGAACUGAGACGUGCAAGA ---(((((((((.((((((((((..((((.........))))....))))).)))))..))))...(((..-((--------........))..))))))))... ( -31.60, z-score = -1.75, R) >droAna3.scaffold_12911 4781028 75 - 5364042 ---UGCAGGACAAUAGCCGAUGUGCGCCGGAAGAAUAAUGGCAUAAACAUUGGACCC---UAUUGUUAAGGAAC-----UGCAAGA------------------- ---((((((((((((((((((((..((((.........))))....)))))))...)---)))))))......)-----))))...------------------- ( -24.30, z-score = -3.62, R) >dp4.chr2 1930985 96 - 30794189 ---GGCAGGACAAUAGCCGAGGUGCGCCGGAAGAAUAAUGGCAUAAACAUCGAACCC-AUUGUCCUGGCGGAGC-----AGCCGAGCGGAGGGAAAACUGCAAGA ---.((((.....(..(((.(((((.(((..((.(((((((..............))-))))).))..))).))-----).))...)))..).....)))).... ( -30.74, z-score = -1.33, R) >droPer1.super_7 2095067 96 - 4445127 ---GGCAGGACAAUAGCCGAGGUGCGCCGGAAGAAUAAUGGCAUAAACAUCGAGCCC-AUUGUCCUGGCGGAGC-----AGCCGAGCGGAGGGAAAACUGCAAGA ---(.(((((((((.(((((.((..((((.........))))....)).))).))..-))))))))).)((...-----..))..((((........)))).... ( -31.60, z-score = -1.19, R) >droMoj3.scaffold_6540 26503171 81 - 34148556 -----------CACAAUGGCUAUGCGCCGGAAGAAUAAUGGCAUUAAUAC--------AUUGAGCAGCAGCAGC-----AGCAGCAUUGUCAGCGCAUUGCAAGA -----------.......((.(((((((....).....(((((.......--------.....((....)).((-----....))..))))))))))).)).... ( -20.70, z-score = 0.41, R) >consensus ___CGCACACAAUGAGCCGAUGUGCGCCGGAAGAAUAAUGGCAUAAACAUUGGGUCCUUUUGUCCUGUCGAAGG______G_AAGGGGAACUGAGACGUGCAAGA ....((((.........((((((..((((.........))))....))))))..((.............))..........................)))).... ( -9.30 = -9.92 + 0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:55 2011