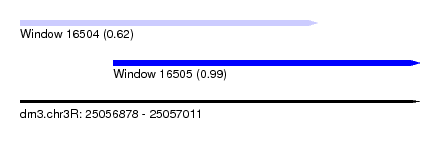
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,056,878 – 25,057,011 |
| Length | 133 |
| Max. P | 0.992889 |

| Location | 25,056,878 – 25,056,977 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 64.56 |
| Shannon entropy | 0.77821 |
| G+C content | 0.58588 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -11.47 |
| Energy contribution | -11.66 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.618751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
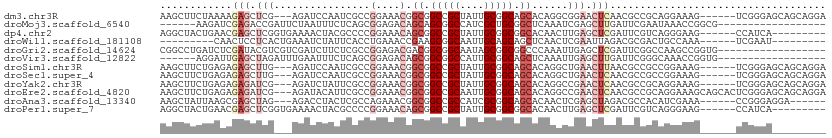
>dm3.chr3R 25056878 99 + 27905053 ---CCCUGGAGGACUCAAAAGAACGCGGGGCAACAAGCUUCUAAAAGAGCU---CGAGAUCCAAUCGCCGGAAACGGCGGCCGCUAUUGCGGCAGCACAGGCGGA ---(((((..((((((...((((.((..(....)..))))))....(....---)))).)))...(((((....)))))(((((....)))))....)))).).. ( -42.30, z-score = -3.15, R) >droMoj3.scaffold_6540 26470595 99 + 34148556 CAACGUCAGCGGUAGAAAUCGCAAGCGUCGU------UCUAAGAUCGAGACCGAUUCUAAUUUCUCAGCGGAGACAGCAGCGGCCAUCGCUGCGGCUCAAAUCGA ..((((..(((((....)))))..))))...------.....(((.(((.((..........((((....))))..((((((.....)))))))))))..))).. ( -29.90, z-score = -0.48, R) >dp4.chr2 1897691 102 + 30794189 ---UGCCGGUGGACGGAAUCGCACGCGACGCGGCAGGCUACUGAACGAGCUCGGUGAAAACUACGCCCCGGAAACAGCGGCCGCUAUUGCGGCGGCACAACUUGA ---..((((.((.((......(((.(((.((..(((....))).....)))))))).......)))))))).....((.(((((....))))).))......... ( -39.42, z-score = -0.78, R) >droWil1.scaffold_181108 1751082 90 + 4707319 ---UAUUCGAGG---CAUUCGAAAGCGAAGAAACCAACUCCUCA------C---UGAAAUCUAUUCACCUGAAACCGAAGCGGCAAUUGCAGCAGCUCAACUCGA ---...(((((.---..((((..((.((.((.......)).)).------)---)........(((....)))..))))((.((....)).)).......))))) ( -15.20, z-score = 0.39, R) >droGri2.scaffold_14624 3268585 102 + 4233967 ---CGUCGGUUUUAGGAAUCGCAAGCGUCGCAACCGGCCUGAUCUCGAUACGUCGUCGAUCUUCUCGCCGGAGACGACGGCGGCAAUAGCGGCGGCCCAAAUUGA ---..(((((((..((...(....)((((((.....(((((....))....(((((((.(((((.....)))))))))))))))....))))))..))))))))) ( -34.80, z-score = 0.56, R) >droVir3.scaffold_12822 2349893 96 + 4096053 ---CGUCGGCGGUCGUAACCGAAAGCGUCGCAA------CAGGAUUGAGCUAGAUUUGAAUUUCUCAGCGGAGACAGCGGCGGCCAUUGCGGCAGCUCAAAUUGA ---....(((.(((((((.(....)((((((..------.........((((((........))).)))(....).))))))....))))))).)))........ ( -29.50, z-score = 0.21, R) >droSim1.chr3R 24727783 99 + 27517382 ---CGCUGGAGGACUCAAAAGAACGCGGGGCAACAAGCUUCUGAGAGAGCU---UGAGAUCCAAUCGCCGGAAACGGCGGCCGCUAUUGCGGCAGCACAGGCUGA ---..........(((...((((.((..(....)..))))))..)))((((---((.........(((((....)))))(((((....)))))....)))))).. ( -42.50, z-score = -3.01, R) >droSec1.super_4 3919662 99 + 6179234 ---CGCUGGAGGACUCAAAAGAACGCGGGGCAACAAGCUUCUGAGAGAGCU---UGAGAUCCAAUCGCCGGAAACGGCGGCCGCUAUUGCGGCAGCACAGGCUGA ---..........(((...((((.((..(....)..))))))..)))((((---((.........(((((....)))))(((((....)))))....)))))).. ( -42.50, z-score = -3.01, R) >droYak2.chr3R 7391669 99 - 28832112 ---CGCUGGAGGAAUCAAAAGGACGCGGGGCAACAAGCUUCUGAGAGAGAU---CGAGAUCUAUUCGCCGGAAACGGCGGCCGCUAUUGCGGCAGCACAGGCCGA ---.(((.((....((...((((.((..(....)..))))))..))....)---)..........(((((....)))))(((((....))))))))......... ( -36.00, z-score = -1.31, R) >droEre2.scaffold_4820 7496533 99 - 10470090 ---CGCUGGAGGACUCAAAAGGACGCGCGGCAACAAGCUUCUGAGAGAGAU---CGAGAUACAUUCGCCGGAAACGGCGGCCGCAAUUGCGGCAGCACAGGCCGA ---.(((....(((((....(((.((..(....)..)).)))....))).)---)..........(((((....)))))(((((....))))))))......... ( -36.70, z-score = -1.71, R) >droAna3.scaffold_13340 9512284 101 - 23697760 -GGAGGAACGGGUCGGGCCAGGACACGCCGGAAUAAGCUAUUAAGCGAGCU---AGAGACCUACUCGCCAGAAACGGCGGCCGCCAUCGCGGCAGCACAACUCGA -.......(((((.(.((.......(((((...(((....))).(((((.(---((....))))))))......)))))(((((....))))).)).).))))). ( -35.40, z-score = -0.89, R) >droPer1.super_7 2061226 102 + 4445127 ---UGCCGGUGGACGUAAUCGCACGCGACGCGGCAGGCUACUGAACGAGCUCGGUGAAAACUACGCCCCGGAAACAGCGGCCGCUAUUGCGGCGGCACAACUUGA ---..((((.((.((((....(((.(((.((..(((....))).....)))))))).....)))))))))).....((.(((((....))))).))......... ( -41.50, z-score = -1.39, R) >consensus ___CGCUGGAGGACGCAAAAGAACGCGGCGCAACAAGCUUCUGAGAGAGCU___UGAGAUCUACUCGCCGGAAACGGCGGCCGCUAUUGCGGCAGCACAAGUUGA ....................................................................(((.....((.(((((....))))).)).....))). (-11.47 = -11.66 + 0.19)
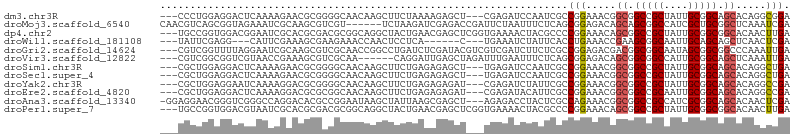
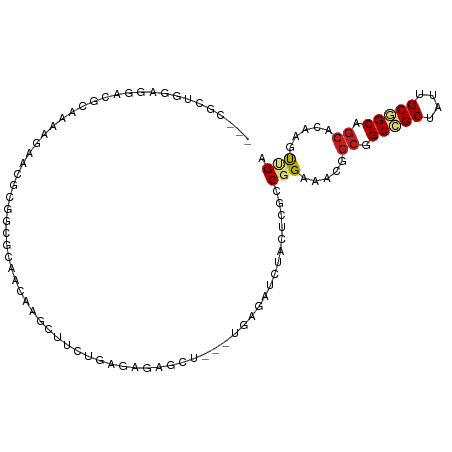
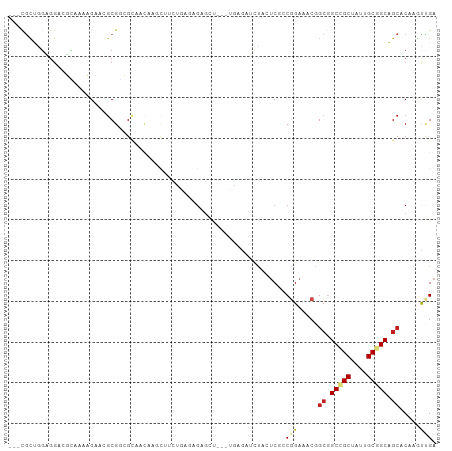
| Location | 25,056,909 – 25,057,011 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 59.60 |
| Shannon entropy | 0.81370 |
| G+C content | 0.57728 |
| Mean single sequence MFE | -37.60 |
| Consensus MFE | -11.95 |
| Energy contribution | -13.02 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.992889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25056909 102 + 27905053 AAGCUUCUAAAAGAGCUCG---AGAUCCAAUCGCCGGAAACGGCGGCCGCUAUUGCGGCAGCACAGGCGGAACUCAACGCCGCAGGAAAG------UCGGGAGCAGCAGGA ..(((((.....))(((((---((.(((...(((((....)))))(((((....))))).((....))))).)))....((((......)------.)))))))))).... ( -44.00, z-score = -2.95, R) >droMoj3.scaffold_6540 26470629 87 + 34148556 ------AAGAUCGAGACCGAUUCUAAUUUCUCAGCGGAGACAGCAGCGGCCAUCGCUGCGGCUCAAAUCGAGCUUGAUUCGAAUAAACCGGCG------------------ ------..(((((((..(((((......((((....))))..((((((.....))))))......)))))..)))))))..............------------------ ( -25.60, z-score = -0.36, R) >dp4.chr2 1897722 96 + 30794189 AGGCUACUGAACGAGCUCGGUGAAAACUACGCCCCGGAAACAGCGGCCGCUAUUGCGGCGGCACAACUUGAGCUCGAUUCGUCAGGGAAG------CCAUCA--------- .((((.((((.(((((((((((.......))))..(....).((.(((((....))))).)).......))))))).....))))...))------))....--------- ( -43.80, z-score = -4.13, R) >droWil1.scaffold_181108 1751110 87 + 4707319 ---------CAACUCCUCACUGAAAUCUAUUCACCUGAAACCGAAGCGGCAAUUGCAGCAGCUCAACUCGAAUUAGACGCGACUGCCAAA------UCGAAU--------- ---------.......(((.((((.....))))..)))...(((...((((.((((..(((.((.....)).)).)..)))).))))...------)))...--------- ( -12.70, z-score = 0.27, R) >droGri2.scaffold_14624 3268616 93 + 4233967 CGGCCUGAUCUCGAUACGUCGUCGAUCUUCUCGCCGGAGACGACGGCGGCAAUAGCGGCGGCCCAAAUUGAGCUCGAUUCGGCCAAGCCGGUG------------------ ..(((...........((((((((.(((((.....)))))))))))))((....))(((((((..(((((....))))).))))..)))))).------------------ ( -37.10, z-score = -0.66, R) >droVir3.scaffold_12822 2349924 87 + 4096053 ------AGGAUUGAGCUAGAUUUGAAUUUCUCAGCGGAGACAGCGGCGGCCAUUGCGGCAGCUCAAAUUGAGCUUGAUUCGGGCAAACCGGUG------------------ ------.((((..((((.(((((((...((((....))))..((.((.((....)).)).))))))))).))))..))))((.....))....------------------ ( -31.90, z-score = -2.04, R) >droSim1.chr3R 24727814 102 + 27517382 AAGCUUCUGAGAGAGCUUG---AGAUCCAAUCGCCGGAAACGGCGGCCGCUAUUGCGGCAGCACAGGCUGAACUUAACGCCGCCGGAAAG------UCGGGAGCAGCAGGA ..((((((((...((((((---.........(((((....)))))(((((....)))))....))))))...(((..((....))..)))------))))))))....... ( -44.70, z-score = -2.55, R) >droSec1.super_4 3919693 102 + 6179234 AAGCUUCUGAGAGAGCUUG---AGAUCCAAUCGCCGGAAACGGCGGCCGCUAUUGCGGCAGCACAGGCUGAACUCAACGCCGCCGGAAAG------UCGGGAGCAGCAGGA ..(((((((((((((((((---.........(((((....)))))(((((....)))))....))))))...)))..((....)).....------))))))))....... ( -44.60, z-score = -2.27, R) >droYak2.chr3R 7391700 102 - 28832112 AAGCUUCUGAGAGAGAUCG---AGAUCUAUUCGCCGGAAACGGCGGCCGCUAUUGCGGCAGCACAGGCCGAACUCAACGCCGCAGGAAAG------UCGGGAGCAGCAGGA ..((((((((..(((.(((---.(.(((...(((((....)))))(((((....))))).....))))))).)))....((...))....------))))))))....... ( -42.00, z-score = -2.19, R) >droEre2.scaffold_4820 7496564 108 - 10470090 AAGCUUCUGAGAGAGAUCG---AGAUACAUUCGCCGGAAACGGCGGCCGCAAUUGCGGCAGCACAGGCCGAACUCAACGCCGCAGGAAAGCAGCACUCGGGAGCAGCAGGA ..(((((((((.(((.(((---.........(((((....)))))(((((....))))).........))).)))...((.((......)).)).)))))))))....... ( -49.70, z-score = -4.72, R) >droAna3.scaffold_13340 9512317 96 - 23697760 AAGCUAUUAAGCGAGCUAG---AGACCUACUCGCCAGAAACGGCGGCCGCCAUCGCGGCAGCACAACUCGAGCUAGACGCCACAUCGAAA------CCGGGAGGA------ ..........(((((.(((---....)))))))).......(((((((((....)))))(((.(.....).)))...)))).........------((....)).------ ( -31.30, z-score = -1.23, R) >droPer1.super_7 2061257 96 + 4445127 AGGCUACUGAACGAGCUCGGUGAAAACUACGCCCCGGAAACAGCGGCCGCUAUUGCGGCGGCACAACUUGAGCUCGAUUCGUCAGGGAAG------CCAUCA--------- .((((.((((.(((((((((((.......))))..(....).((.(((((....))))).)).......))))))).....))))...))------))....--------- ( -43.80, z-score = -4.13, R) >consensus AAGCUUCUGAGAGAGCUCG___AGAUCUACUCGCCGGAAACGGCGGCCGCUAUUGCGGCAGCACAAGUUGAACUCGACGCCGCAGGAAAG______UCGGGA_________ ............(((.((.................(....).((.(((((....))))).)).......)).))).................................... (-11.95 = -13.02 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:55 2011