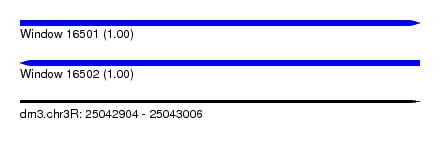
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,042,904 – 25,043,006 |
| Length | 102 |
| Max. P | 0.998860 |

| Location | 25,042,904 – 25,043,006 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.73 |
| Shannon entropy | 0.10543 |
| G+C content | 0.42500 |
| Mean single sequence MFE | -36.37 |
| Consensus MFE | -35.14 |
| Energy contribution | -35.14 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.04 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.998860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
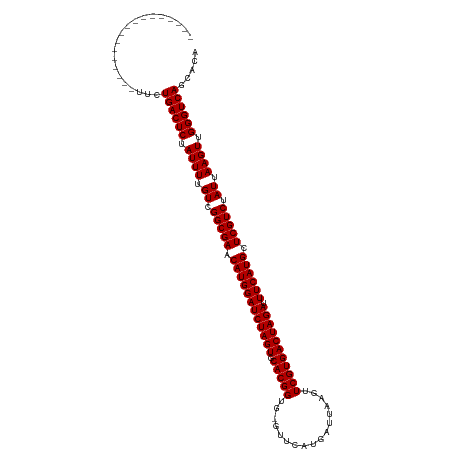
>dm3.chr3R 25042904 102 + 27905053 -----------------UUCUGACUCUAUUUUGUCGGCGAACAUGGAUCUAGUGCACGGUG-GUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGGUCAGCACA -----------------..(((((((.((((.((.(((((.((((((((((((.(((((..-..............))))))))))).)))))).))))).)).)))).))))))).... ( -37.39, z-score = -4.28, R) >droSim1.chr3R 24713800 102 + 27517382 -----------------UUCUGACUCUAUUUUGUCGGCGAACAUGGAUCUAGUGCACGGUG-GUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGGUCAGCACA -----------------..(((((((.((((.((.(((((.((((((((((((.(((((..-..............))))))))))).)))))).))))).)).)))).))))))).... ( -37.39, z-score = -4.28, R) >droSec1.super_4 3905697 102 + 6179234 -----------------UUCUGACUCUAUUUUGUCGGCGAACAUGGAUCUAGUGCACGGUG-GUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGGUCAGCACA -----------------..(((((((.((((.((.(((((.((((((((((((.(((((..-..............))))))))))).)))))).))))).)).)))).))))))).... ( -37.39, z-score = -4.28, R) >droYak2.chr3R 7377586 102 - 28832112 -----------------UUCUGACUCUAUUUUGUCGGCGAACAUGGAUCUAGUGCACGGUG-GUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGGUCAGCACU -----------------..(((((((.((((.((.(((((.((((((((((((.(((((..-..............))))))))))).)))))).))))).)).)))).))))))).... ( -37.39, z-score = -4.32, R) >droEre2.scaffold_4820 7482567 102 - 10470090 -----------------UUCUGACUCUAUUUUGUCGGCGAACAUGGAUCUAGUGCACGGUG-GUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGGUCAGCACU -----------------..(((((((.((((.((.(((((.((((((((((((.(((((..-..............))))))))))).)))))).))))).)).)))).))))))).... ( -37.39, z-score = -4.32, R) >droAna3.scaffold_13340 9498697 102 - 23697760 -----------------UUCUGACUCUAUUUUGUCGGCGAACAUGGAUCUAGUGCACGGUG-UUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGGUCAGCACA -----------------..(((((((.((((.((.(((((.((((((((((((.(((((..-((........))..))))))))))).)))))).))))).)).)))).))))))).... ( -37.40, z-score = -4.50, R) >droPer1.super_7 2047033 102 + 4445127 -----------------CUCUGACUCUAUUUUGUCGGCGAACAUGGAUCUAGUGCACGGUU-GUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGGUCAACACA -----------------...((((((.((((.((.(((((.((((((((((((.(((((..-..............))))))))))).)))))).))))).)).)))).))))))..... ( -35.19, z-score = -4.02, R) >dp4.chr2 1883620 102 + 30794189 -----------------CUCUGACUCUAUUUUGUCGGCGAACAUGGAUCUAGUGCACGGUU-GUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGGUCAACACA -----------------...((((((.((((.((.(((((.((((((((((((.(((((..-..............))))))))))).)))))).))))).)).)))).))))))..... ( -35.19, z-score = -4.02, R) >droWil1.scaffold_181108 1735992 120 + 4707319 UUCUAUGUGGCAGCGUCUCUUGACUCUAUUUUGUCGGCGAACAUGGAUCUAGUGCACGGUUUGUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGGUCAGCACA .....((((..........(((((((.((((.((.(((((.((((((((((((.(((((.................))))))))))).)))))).))))).)).)))).))))))))))) ( -36.13, z-score = -2.09, R) >droVir3.scaffold_12822 2333413 102 + 4096053 -----------------CUCUGACUCUAUUUUGUCGGCGAACAUGGAUCUAGUGCACGGUU-GUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGGUCAACACA -----------------...((((((.((((.((.(((((.((((((((((((.(((((..-..............))))))))))).)))))).))))).)).)))).))))))..... ( -35.19, z-score = -4.02, R) >droMoj3.scaffold_6540 26449283 102 + 34148556 -----------------CUCUGACUCUAUUUUGUCGGCGAACAUGGAUCUAGUGCACGGUU-GUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGGUCAAUACA -----------------...((((((.((((.((.(((((.((((((((((((.(((((..-..............))))))))))).)))))).))))).)).)))).))))))..... ( -35.19, z-score = -4.25, R) >droGri2.scaffold_14624 3251573 102 + 4233967 -----------------UUCUGACUCUAUUUUGUCGGCGAACAUGGAUCUAGUGCACGGUU-GUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGGUCAACACA -----------------...((((((.((((.((.(((((.((((((((((((.(((((..-..............))))))))))).)))))).))))).)).)))).))))))..... ( -35.19, z-score = -4.10, R) >consensus _________________UUCUGACUCUAUUUUGUCGGCGAACAUGGAUCUAGUGCACGGUG_GUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGGUCAGCACA ....................((((((.((((.((.(((((.((((((((((((.(((((.................))))))))))).)))))).))))).)).)))).))))))..... (-35.14 = -35.14 + 0.00)

| Location | 25,042,904 – 25,043,006 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.73 |
| Shannon entropy | 0.10543 |
| G+C content | 0.42500 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -27.21 |
| Energy contribution | -27.09 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.74 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.48 |
| SVM RNA-class probability | 0.998763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
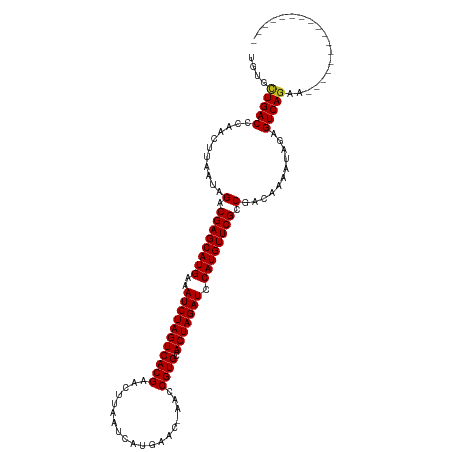
>dm3.chr3R 25042904 102 - 27905053 UGUGCUGACCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAAC-CACCGUGCACUAGAUCCAUGUUCGCCGACAAAAUAGAGUCAGAA----------------- ....(((((...........(.((((((((..(((((((((((...............-...)))).))))))).)))))))).)...........)))))..----------------- ( -28.42, z-score = -4.19, R) >droSim1.chr3R 24713800 102 - 27517382 UGUGCUGACCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAAC-CACCGUGCACUAGAUCCAUGUUCGCCGACAAAAUAGAGUCAGAA----------------- ....(((((...........(.((((((((..(((((((((((...............-...)))).))))))).)))))))).)...........)))))..----------------- ( -28.42, z-score = -4.19, R) >droSec1.super_4 3905697 102 - 6179234 UGUGCUGACCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAAC-CACCGUGCACUAGAUCCAUGUUCGCCGACAAAAUAGAGUCAGAA----------------- ....(((((...........(.((((((((..(((((((((((...............-...)))).))))))).)))))))).)...........)))))..----------------- ( -28.42, z-score = -4.19, R) >droYak2.chr3R 7377586 102 + 28832112 AGUGCUGACCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAAC-CACCGUGCACUAGAUCCAUGUUCGCCGACAAAAUAGAGUCAGAA----------------- ....(((((...........(.((((((((..(((((((((((...............-...)))).))))))).)))))))).)...........)))))..----------------- ( -28.42, z-score = -4.21, R) >droEre2.scaffold_4820 7482567 102 + 10470090 AGUGCUGACCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAAC-CACCGUGCACUAGAUCCAUGUUCGCCGACAAAAUAGAGUCAGAA----------------- ....(((((...........(.((((((((..(((((((((((...............-...)))).))))))).)))))))).)...........)))))..----------------- ( -28.42, z-score = -4.21, R) >droAna3.scaffold_13340 9498697 102 + 23697760 UGUGCUGACCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAAA-CACCGUGCACUAGAUCCAUGUUCGCCGACAAAAUAGAGUCAGAA----------------- ....(((((...........(.((((((((..(((((((((((...............-...)))).))))))).)))))))).)...........)))))..----------------- ( -28.42, z-score = -4.12, R) >droPer1.super_7 2047033 102 - 4445127 UGUGUUGACCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAAC-AACCGUGCACUAGAUCCAUGUUCGCCGACAAAAUAGAGUCAGAG----------------- ....(((((...........(.((((((((..(((((((((((...............-...)))).))))))).)))))))).)...........)))))..----------------- ( -26.12, z-score = -3.26, R) >dp4.chr2 1883620 102 - 30794189 UGUGUUGACCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAAC-AACCGUGCACUAGAUCCAUGUUCGCCGACAAAAUAGAGUCAGAG----------------- ....(((((...........(.((((((((..(((((((((((...............-...)))).))))))).)))))))).)...........)))))..----------------- ( -26.12, z-score = -3.26, R) >droWil1.scaffold_181108 1735992 120 - 4707319 UGUGCUGACCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAACAAACCGUGCACUAGAUCCAUGUUCGCCGACAAAAUAGAGUCAAGAGACGCUGCCACAUAGAA ((((................(.((((((((..(((((((((((...................)))).))))))).)))))))).).......(((.(((....))).))).))))..... ( -29.01, z-score = -2.90, R) >droVir3.scaffold_12822 2333413 102 - 4096053 UGUGUUGACCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAAC-AACCGUGCACUAGAUCCAUGUUCGCCGACAAAAUAGAGUCAGAG----------------- ....(((((...........(.((((((((..(((((((((((...............-...)))).))))))).)))))))).)...........)))))..----------------- ( -26.12, z-score = -3.26, R) >droMoj3.scaffold_6540 26449283 102 - 34148556 UGUAUUGACCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAAC-AACCGUGCACUAGAUCCAUGUUCGCCGACAAAAUAGAGUCAGAG----------------- ....(((((...........(.((((((((..(((((((((((...............-...)))).))))))).)))))))).)...........)))))..----------------- ( -26.22, z-score = -3.84, R) >droGri2.scaffold_14624 3251573 102 - 4233967 UGUGUUGACCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAAC-AACCGUGCACUAGAUCCAUGUUCGCCGACAAAAUAGAGUCAGAA----------------- ....(((((...........(.((((((((..(((((((((((...............-...)))).))))))).)))))))).)...........)))))..----------------- ( -26.12, z-score = -3.29, R) >consensus UGUGCUGACCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAAC_AACCGUGCACUAGAUCCAUGUUCGCCGACAAAAUAGAGUCAGAA_________________ ....(((((...........(.((((((((..(((((((((((...................)))).))))))).)))))))).)...........)))))................... (-27.21 = -27.09 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:52 2011