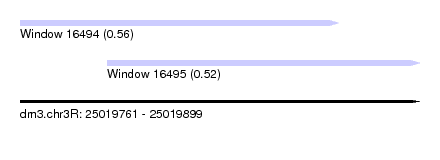
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,019,761 – 25,019,899 |
| Length | 138 |
| Max. P | 0.563153 |

| Location | 25,019,761 – 25,019,871 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.17 |
| Shannon entropy | 0.46683 |
| G+C content | 0.36316 |
| Mean single sequence MFE | -15.98 |
| Consensus MFE | -8.98 |
| Energy contribution | -9.02 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.563153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25019761 110 + 27905053 AAUCAUACUUAUAUACAUA----------CGCAGCAAGUCACAUUUCACGAAUGACAAAUUCCAUUGCCUACUUUUAGGCACUACCACAACUUUAGGCACCACAUUGCUUUUAAAUAGCC ...................----------.((...((((..........((((.....))))...((((((....))))))........))))...))........(((.......))). ( -13.80, z-score = -1.18, R) >droEre2.scaffold_4820 7459347 107 - 10470090 AAUCGUCCAUACUAAUAUGGUUUUUAUACAGCUCCCAGUUGCAUUUCCCGAUUGAAAUAUUCCGUUGCCUACUUUUAGGCAGUACUAC---------CAC----UUGCCUUCAAAUAUCU (((((.(((((....)))))........((((.....)))).......)))))...........(((((((....)))))))......---------...----................ ( -16.20, z-score = -1.05, R) >droYak2.chr3R 7352969 116 - 28832112 AUUCAUCAAUUUUAAGAUGGCUUA-ACACCGCUCCAUGUCACAUUUCCCGAAUGAAAUAUUUCAUUGCCUACUUUUAGGCACUACUACU---UAAAGCACAAGAUUGCUUUCAAAUACCU ...((((........))))((...-.....))..................((((((....))))))(((((....))))).........---.((((((......))))))......... ( -16.40, z-score = -1.27, R) >droSec1.super_4 3882916 109 + 6179234 AAUCAUCCAUAUUAAUAU-----------CGCAGCAAGUCACAUUUCCCAAAUGAAAAAUUCCAUUGCCUACUUUUAGGCACUAAUACAACUUUGGGCUCCAAAUUGCUUUCAAAUACAG ..................-----------...(((((.........((((((((........)).((((((....))))))..........)))))).......)))))........... ( -16.29, z-score = -1.76, R) >droSim1.chr3R 24690759 109 + 27517382 AAUCAUAAAUAUUAAUAU-----------CGCAGCAAGUCACAUUUCCCGAAUGAAAAAUUCCAUUGCCUACUUUUAGGCACCAAUACAACUUUGGGCACCAAAUUGCUUUCAAAUGCAG ..................-----------.(((((((.........(((((((.....))))...((((((....)))))).............))).......)))).......))).. ( -17.20, z-score = -0.97, R) >consensus AAUCAUCCAUAUUAAUAU___________CGCAGCAAGUCACAUUUCCCGAAUGAAAAAUUCCAUUGCCUACUUUUAGGCACUACUACAACUUUAGGCACCAAAUUGCUUUCAAAUACCG ..............................((.........(((((...)))))...........((((((....)))))).........................))............ ( -8.98 = -9.02 + 0.04)


| Location | 25,019,791 – 25,019,899 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.02 |
| Shannon entropy | 0.45041 |
| G+C content | 0.34028 |
| Mean single sequence MFE | -15.91 |
| Consensus MFE | -8.06 |
| Energy contribution | -9.18 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.515316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25019791 108 + 27905053 ACAUUUCACGAAUGACAAAUUCCAUUGCCUACUUUUAGGCACUACCACAACUUUAGGCACCACAUUGCUUUUAAAUAGC------------CAUAACUUUAAGAUCUUUUAUAGAUUUCU .........((((.....))))...((((((....))))))..............(((...................))------------).........((((((.....)))))).. ( -13.81, z-score = -1.85, R) >droEre2.scaffold_4820 7459387 86 - 10470090 GCAUUUCCCGAUUGAAAUAUUCCGUUGCCUACUUUUAGGCAGUACUACCACUU-------------GCCUUCAAAUA---------------------UCUAUAUCUUGUGUAGAUUUGG .......(((((((((.........((((((....))))))(((........)-------------)).)))))..(---------------------(((((((...)))))))))))) ( -15.40, z-score = -1.52, R) >droYak2.chr3R 7353008 117 - 28832112 ACAUUUCCCGAAUGAAAUAUUUCAUUGCCUACUUUUAGGCACUACUA---CUUAAAGCACAAGAUUGCUUUCAAAUACCUACAUACAUAUACAUAUCUACAAGUUCUUGUAUAAAUUUCU ..........((((((....))))))(((((....))))).......---...((((((......))))))................((((((..............))))))....... ( -15.94, z-score = -2.36, R) >droSec1.super_4 3882945 108 + 6179234 ACAUUUCCCAAAUGAAAAAUUCCAUUGCCUACUUUUAGGCACUAAUACAACUUUGGGCUCCAAAUUGCUUUCAAAUACA------------GAGAACUUCAAGAUCUUGUAUAGAUUUCU ............(((((........((((((....))))))......(((.(((((...)))))))).))))).(((((------------.(((.(.....).))))))))........ ( -16.20, z-score = -0.60, R) >droSim1.chr3R 24690788 108 + 27517382 ACAUUUCCCGAAUGAAAAAUUCCAUUGCCUACUUUUAGGCACCAAUACAACUUUGGGCACCAAAUUGCUUUCAAAUGCA------------GAUAACUUCAAGAUCUUGUAUAGAUUUCU ......(((((((.....))))...((((((....)))))).............))).......((((........)))------------).........((((((.....)))))).. ( -18.20, z-score = -1.12, R) >consensus ACAUUUCCCGAAUGAAAAAUUCCAUUGCCUACUUUUAGGCACUACUACAACUUUAGGCACCAAAUUGCUUUCAAAUACA____________CAUAACUUCAAGAUCUUGUAUAGAUUUCU ............((((.........((((((....))))))..............(((........)))))))............................((((((.....)))))).. ( -8.06 = -9.18 + 1.12)

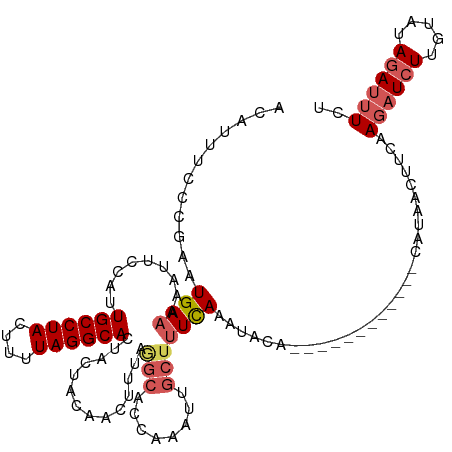

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:46 2011