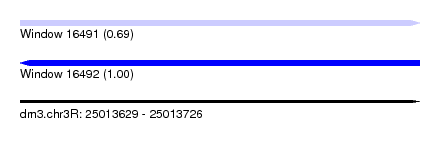
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,013,629 – 25,013,726 |
| Length | 97 |
| Max. P | 0.998213 |

| Location | 25,013,629 – 25,013,726 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 93.24 |
| Shannon entropy | 0.11738 |
| G+C content | 0.37445 |
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -17.34 |
| Energy contribution | -17.54 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.691561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
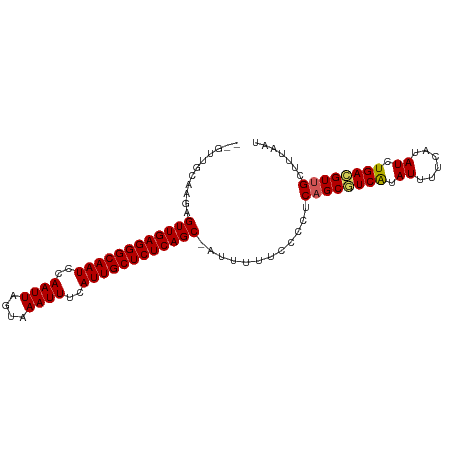
>dm3.chr3R 25013629 97 + 27905053 AUUAAAGCAACGUCAGAUAUGAAAAUCUGAUGCUGAGGGGAAAAAU-GCUGAGAGCAAUGAAAUUUACUAAUUGGAUUGCCCUCAACUCGUGCAACUC ......(((.((((((((......)))))))).((((((......(-((.....)))....((((((.....)))))).)))))).....)))..... ( -24.80, z-score = -2.48, R) >droEre2.scaffold_4820 7453429 95 - 10470090 AUUAAAGCAACGUCAGAUAUGAAAAUACGACGCUGAAGGUAAAAAU-GCUGAGAGCAAUGAAAUUUGCUAAUUGGAUUGCCCUCAACUCUUGCAAA-- ......((((((((.(.(((....)))))))).(((.(((((....-.(..(.(((((......)))))..)..).))))).)))....))))...-- ( -20.80, z-score = -1.72, R) >droYak2.chr3R 7346617 96 - 28832112 AUUAAAGCAACAUCAGAUAUGAAAAUAUGAUGCUGAAGGGAAAAAUUGCUGAGAGCAAUGAAAUUUGCUAAUUGGAUUGCCCUCAACUCUUGCAAG-- ......(((((((((..(((....)))))))).(((.(((...((((......(((((......))))).....)))).))))))....))))...-- ( -21.00, z-score = -1.28, R) >droSec1.super_4 3876696 95 + 6179234 AUUAAAGCCACGUCAGAUAUGAAAAUAUGACGCUGAGGUGAAAAAU-GCUGAGAGCAAUGAAAUUUACUAAUUGGAUUGCCCUCAACUCUUGCAAC-- ......(((((.((((((((....))))....)))).)))......-..((((.(((((..((((....))))..))))).))))......))...-- ( -22.70, z-score = -2.80, R) >droSim1.chr3R 24684572 95 + 27517382 AUUAAAGCAACGUCAGAUAUGAAAAUAUGACGCUGAGGGGAAAAAU-GCUGAGAGCAAUGAAAUUUACUAAUUGGAUUGCCCUCAACUCUUGCAAC-- ......(((((((((..(((....)))))))).((((((......(-((.....)))....((((((.....)))))).))))))....))))...-- ( -22.30, z-score = -2.43, R) >consensus AUUAAAGCAACGUCAGAUAUGAAAAUAUGACGCUGAGGGGAAAAAU_GCUGAGAGCAAUGAAAUUUACUAAUUGGAUUGCCCUCAACUCUUGCAAC__ ......(((((((((.((......)).))))).................((((.(((((..((((....))))..))))).))))....))))..... (-17.34 = -17.54 + 0.20)
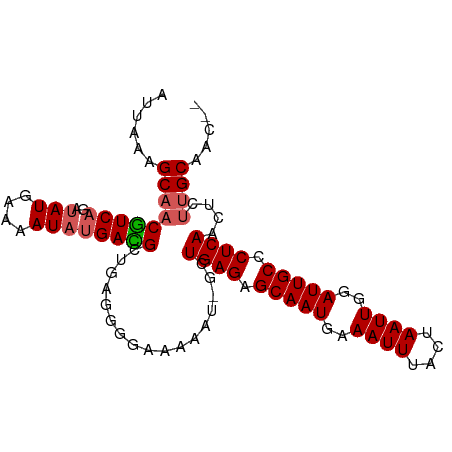
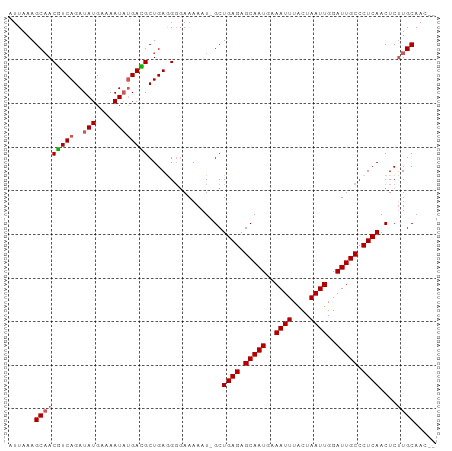
| Location | 25,013,629 – 25,013,726 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 93.24 |
| Shannon entropy | 0.11738 |
| G+C content | 0.37445 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -24.06 |
| Energy contribution | -24.06 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.87 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25013629 97 - 27905053 GAGUUGCACGAGUUGAGGGCAAUCCAAUUAGUAAAUUUCAUUGCUCUCAGC-AUUUUUCCCCUCAGCAUCAGAUUUUCAUAUCUGACGUUGCUUUAAU .....(((((.((((((((((((..((((....))))..))))))))))))-................((((((......)))))))).)))...... ( -28.00, z-score = -3.94, R) >droEre2.scaffold_4820 7453429 95 + 10470090 --UUUGCAAGAGUUGAGGGCAAUCCAAUUAGCAAAUUUCAUUGCUCUCAGC-AUUUUUACCUUCAGCGUCGUAUUUUCAUAUCUGACGUUGCUUUAAU --.........((((((((((((..((((....))))..))))))))))))-...........((((((((.((......)).))))))))....... ( -26.60, z-score = -3.91, R) >droYak2.chr3R 7346617 96 + 28832112 --CUUGCAAGAGUUGAGGGCAAUCCAAUUAGCAAAUUUCAUUGCUCUCAGCAAUUUUUCCCUUCAGCAUCAUAUUUUCAUAUCUGAUGUUGCUUUAAU --.........((((((((((((..((((....))))..))))))))))))............((((((((.((......)).))))))))....... ( -25.90, z-score = -3.21, R) >droSec1.super_4 3876696 95 - 6179234 --GUUGCAAGAGUUGAGGGCAAUCCAAUUAGUAAAUUUCAUUGCUCUCAGC-AUUUUUCACCUCAGCGUCAUAUUUUCAUAUCUGACGUGGCUUUAAU --...((....((((((((((((..((((....))))..))))))))))))-.............((((((.((......)).)))))).))...... ( -26.30, z-score = -3.71, R) >droSim1.chr3R 24684572 95 - 27517382 --GUUGCAAGAGUUGAGGGCAAUCCAAUUAGUAAAUUUCAUUGCUCUCAGC-AUUUUUCCCCUCAGCGUCAUAUUUUCAUAUCUGACGUUGCUUUAAU --.........((((((((((((..((((....))))..))))))))))))-...........((((((((.((......)).))))))))....... ( -27.30, z-score = -4.60, R) >consensus __GUUGCAAGAGUUGAGGGCAAUCCAAUUAGUAAAUUUCAUUGCUCUCAGC_AUUUUUCCCCUCAGCGUCAUAUUUUCAUAUCUGACGUUGCUUUAAU ...........((((((((((((..((((....))))..))))))))))))............((((((((.((......)).))))))))....... (-24.06 = -24.06 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:44 2011