| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,501 – 19,654 |
| Length | 153 |
| Max. P | 0.992602 |

| Location | 19,501 – 19,654 |
|---|---|
| Length | 153 |
| Sequences | 5 |
| Columns | 159 |
| Reading direction | forward |
| Mean pairwise identity | 67.33 |
| Shannon entropy | 0.58789 |
| G+C content | 0.42192 |
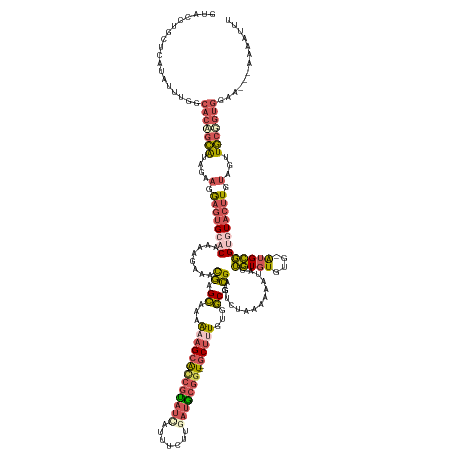
| Mean single sequence MFE | -46.46 |
| Consensus MFE | -23.52 |
| Energy contribution | -25.48 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.992602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
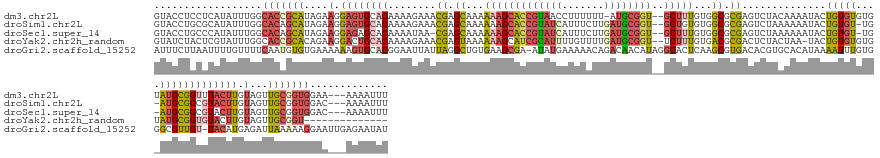
>dm3.chr2L 19501 153 + 23011544 GUACCUCCUCAUAUUUGGCACCGCAUAGAAGGAGUGCACAAAAGAAACGAGCAAAAAAGCACCGUAACCUUUUUU-AUGCGGU--GCUUUGUGGCGCGAGUCUACAAAAUACUGUGUGUGUAUGCGGUUUACUUGUAGUUGCGGUGGAA---AAAAUUU ...((.(((((((...((((((((((((((((.((((.....................)))).....)))))..)-)))))))--))).))))).((((..((((((...(((((((....)))))))....)))))))))))).))..---....... ( -47.80, z-score = -2.04, R) >droSim1.chr2L 23159 152 + 22036055 GUACCUGCGCAUAUUUGGCACAGCAUAGAAGGAGUGCACAAAAGAAACGAGCAAAAAAGCACCGUAUCAUUUCUUGAUGCGGU--GCUGUGUGGCGCGAGUCUAAAAAAUACUGUGU-UG-AUGCGGCGUACUUGUAGUUGCGGUGGAC---AAAAUUU .((((..((((...(..(((((((((.......)))).....(((..((.((.....((((((((((((.....)))))))))--))).....)).))..))).........)))))-..-)))))(((.((.....)))))))))...---....... ( -51.50, z-score = -2.25, R) >droSec1.super_14 24665 151 + 2068291 GUACCUGCCCAUAUUUGGCACAGCAUAGAAGGAGAGCACAAAAUAA-CGAGCAAAAAAGCACCGUAUCAUUUCUUGAUGCGGU--GCUUUGUGGCGCGAGUCUAAAAAAUACUGUGU-UG-AUGCGGCGUACUUGUAGUUGCGGUGGAC---AAAAUUU .((((((((.......))))((((((((..(.......).......-((.((...((((((((((((((.....)))))))))--)))))...)).)).............))))))-))-..(((((.........)))))))))...---....... ( -50.30, z-score = -2.59, R) >droYak2.chr2h_random 1951082 142 + 3774259 GUAUCUACUCGUAUUUGGCACCGCACAGAAGGACUGCACAAAAGAAACGAGUAAAAAAGCAUCGCAUUUUGUUUUGAUGCGGU--UCUUUGUGACGCGACUCUACUAA-UACUGUGUGUGUAUGCGGUGUACUUGUAGUUGCGGU-------------- (((.((((..((.....((((((((..((((((((((((((((.((((((((......)).)))...))).))))).))))))--)))))...(((((((........-....)).))))).))))))))))..)))).)))...-------------- ( -44.50, z-score = -1.66, R) >droGri2.scaffold_15252 38589 157 - 17193109 AUUUCUUAAUUUUGUUUUCAAUGUGUGAAAAAAGUGCACGGAAUUAUUAGGCUGUGAAGCGA-AUAUGAAAAACAGACAACAUAGGCACUCAAGCGUGACACGUGCACAUAAAAUUUGUGGGCGUUGU-UACAUGAGAUUAAAAAGGAAUUGAGAAUAU ..(((((((((((.((((((.(((((.........)))))....((((..(((....))).)-)))))))))................((((...(((((((((.((((.......)))).))).)))-))).))))........)))))))))))... ( -38.20, z-score = -2.00, R) >consensus GUACCUGCUCAUAUUUGGCACAGCAUAGAAGGAGUGCACAAAAGAAACGAGCAAAAAAGCACCGUAUCAUUUCUUGAUGCGGU__GCUUUGUGGCGCGAGUCUAAAAAAUACUGUGUGUG_AUGCGGUGUACUUGUAGUUGCGGUGGAA___AAAAUUU ..................(((((((....(.((((((((........((.((...(((((((((((((.......))))))))..)))))...)).))..............(((((....))))))))))))).)...)))))))............. (-23.52 = -25.48 + 1.96)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:04:54 2011