| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,008,334 – 25,008,431 |
| Length | 97 |
| Max. P | 0.728251 |

| Location | 25,008,334 – 25,008,431 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 57.04 |
| Shannon entropy | 0.77061 |
| G+C content | 0.42043 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -8.91 |
| Energy contribution | -9.24 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.75 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.728251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
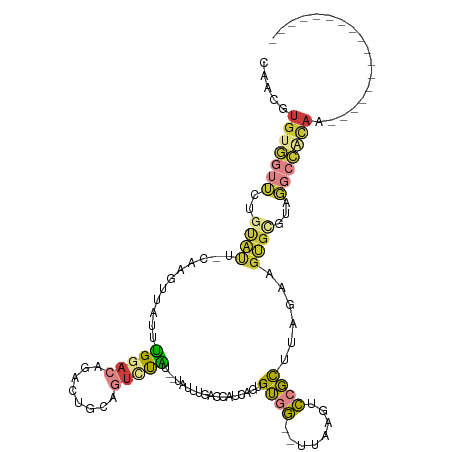
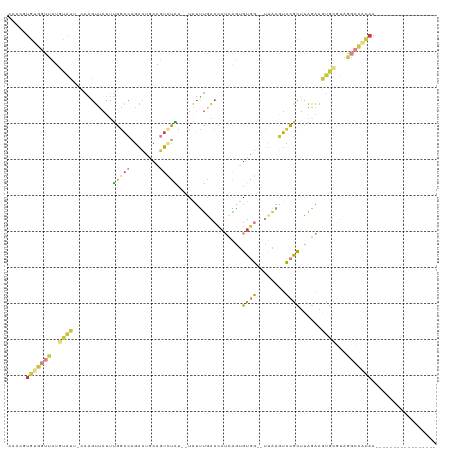
>dm3.chr3R 25008334 97 - 27905053 ---CAUGUGGUUCUGUAUUCCAAGUUAUUUGGACAGACUGCAGUCUAU--UAUUUAACCAUCAGUGUGGUUUUAGGUCCGCUUAGAAGUGUGUCGGCCACAA----------------- ---...((((((..(((.((((((...)))))).((((....))))..--)))..))))))...((((((....((..((((....))))..)).)))))).----------------- ( -25.60, z-score = -1.39, R) >droSim1.chr3R 24680577 99 - 27517382 CAACGUGUGGUUCUGUACU-CAAGUUAUUUGGACAGACUGCAGUCUAU--UAUUUGACCAUCAGUGUGGUUUUAGGUCCGCUCAGAAGUGCGUAGGCCACAA----------------- .....(((((((.((((((-(.(((.....((((((((....))))..--.....((((((....))))))....)))))))..).))))))..))))))).----------------- ( -27.00, z-score = -0.67, R) >droSec1.super_4 3872737 99 - 6179234 CAACGUGUGGUUCUGUACU-CAAGUUAUUUGGACAGACUGCAGUCUAU--UAUUUGACCAUCAGUGUGGUUUUAGGUCCGCUCAGAAGUGCGUAGGCCACAA----------------- .....(((((((.((((((-(.(((.....((((((((....))))..--.....((((((....))))))....)))))))..).))))))..))))))).----------------- ( -27.00, z-score = -0.67, R) >droYak2.chr3R 7342482 97 + 28832112 CAACGUGUGGUUUUGCAUU-CAAGUUAUUUGGACAGACUGCAGUCCAU--UAUUUGACCAACAGUGUGG--UUAAGUCCGCUUAGAAGUGCGUAGGCCACAA----------------- .....((((((((((((((-(((((....(((((........))))).--.(((((((((......)))--))))))..)))).).)))))).)))))))).----------------- ( -31.60, z-score = -2.65, R) >droEre2.scaffold_4820 7448984 99 + 10470090 CAACGUGUGGUUUAGCAUG-CAGGUUAUUUGGACAGACUGCCGUACAUUCUAUUUGACCAUCGAUGUGG--UUAAGUCCGCUUAGAAGUGCGUAGGCCGCAA----------------- .....(((((((((((((.-....(((..((((((((.((.....)).)))..((((((((....))))--)))))))))..)))..)))).))))))))).----------------- ( -32.10, z-score = -1.87, R) >droAna3.scaffold_13340 9465016 78 + 23697760 ---UAUGCAAUUAUUUAU----AGCCCUUGGGGAAGCCGUCAAGUUUUCAUUUCCAACAAUUGUGGUUA--GCCAUUUUGUCUGAAA-------------------------------- ---...............----....(((((.(....).)))))......((((..((((..((((...--.)))).))))..))))-------------------------------- ( -12.70, z-score = 0.62, R) >droGri2.scaffold_14624 3213394 112 - 4233967 --CCGUGAAGGGUUUCGC--AAUGUUAUAAGCAGAAAUUUAAAUAUU---UGAGUGGGAGAAAUUGAAAAAGAGUAUUCUUUUAAGAGCGCUCUCAAUAUAAAAAUGCGACACAUAAAA --((.....))((.((((--(...(((((.......((((((....)---)))))(((((...((..(((((......)))))..))...)))))..)))))...))))).))...... ( -22.80, z-score = -1.58, R) >consensus CAACGUGUGGUUCUGUAUU_CAAGUUAUUUGGACAGACUGCAGUCUAU__UAUUUGACCAUCAGUGUGG__UUAAGUCCGCUUAGAAGUGCGUAGGCCACAA_________________ .....(((((((..((((...........(((((........)))))..................((((........))))......))))...))))))).................. ( -8.91 = -9.24 + 0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:42 2011