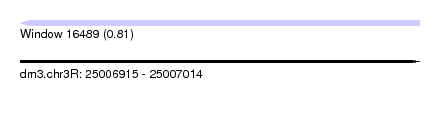
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,006,915 – 25,007,014 |
| Length | 99 |
| Max. P | 0.813242 |

| Location | 25,006,915 – 25,007,014 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.44 |
| Shannon entropy | 0.46980 |
| G+C content | 0.55982 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -14.60 |
| Energy contribution | -13.19 |
| Covariance contribution | -1.41 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.813242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25006915 99 - 27905053 ---CCUUAACGGCC---UGCUCCUGUGCCUGAUUUCCGUU--------------AUUGCC-CUGCUGCCGCCGCUGACGCUCGGCGAUGACGGCGAUAAUUUCUUUGCCGUGAAUGCCUU ---.....(((((.---.((......))...(((.(((((--------------((((((-..((.((....))....))..))))))))))).))).........)))))......... ( -30.30, z-score = -2.53, R) >droSim1.chr3R 24679168 99 - 27517382 ---CCUUAACGGCC---UGCUCCUGUGCCUGAUUUCCGUU--------------AUUGCC-CUGCUGCCGCCGCUGACGCUCGGCGAUGACGGCGAUAAUUUCUUUGCCGUGAAUGCCUU ---.....(((((.---.((......))...(((.(((((--------------((((((-..((.((....))....))..))))))))))).))).........)))))......... ( -30.30, z-score = -2.53, R) >droSec1.super_4 3871343 99 - 6179234 ---CCUUAACGGCC---UGCUCCUGUGCCUGAUUUCCGUU--------------AUUGCC-CUGCUGCCGCCGCUGACGCUCGGCGAUGACGGCGAUAAUUUCUUUGCCGUGAAUGCCUU ---.....(((((.---.((......))...(((.(((((--------------((((((-..((.((....))....))..))))))))))).))).........)))))......... ( -30.30, z-score = -2.53, R) >droYak2.chr3R 7341011 99 + 28832112 ---CCUUAACGGCC---UGCUCCUGUGCCUGAUUUCCGUU--------------AUUGCC-CUGCUGCCGCCGCUGACGCUCGGCGAUGACGGCGAUAAUUUCUUUGCCGUGAAUGCCUU ---.....(((((.---.((......))...(((.(((((--------------((((((-..((.((....))....))..))))))))))).))).........)))))......... ( -30.30, z-score = -2.53, R) >droEre2.scaffold_4820 7447543 99 + 10470090 ---CCUUAACGGCC---UGCUCCUGUGCCUGAUUUCCGUU--------------GUUGCC-CUGCUGCCGCCGCUGACGCUCGGCGAUGACGGCGAUAAUUUCUUUGCCGUGAAUGCCUU ---.....(((((.---.((......))...(((.((((.--------------.(((((-..((.((....))....))..)))))..)))).))).........)))))......... ( -29.40, z-score = -1.96, R) >droAna3.scaffold_13340 9463376 102 + 23697760 ---UCUCAACUCGUCCCUCGUUCUGCUACUGAUAUCCGUU--------------AUUGCC-CUAAUGCCACCGCUGACGCUGGGUGAUGACGGUGAUAAUUUCUUUGCCGUGAAUGCCUU ---...............(((((.((....((...(((((--------------((..((-(....((....)).......)))..)))))))........))...))...))))).... ( -22.50, z-score = -1.10, R) >dp4.chr2 1849291 99 - 30794189 ---CCUCAGCACGU---UUGUCCUGUGCCUGAUUUCCGUC--------------AUUGCC-CUGCUGCCGCCUCUAACGCUCGGCGAUGACGGUGAUAAUUUCUUUGCCGUAAAUGCCUU ---..((((((((.---......)))).)))).....(((--------------((((((-..((.............))..)))))))))((..(........)..))........... ( -27.82, z-score = -2.62, R) >droPer1.super_7 2012917 99 - 4445127 ---CCUCAGCACGU---UUGUCCUGUGCCUGAUUUCCGUC--------------AUUGCC-CUGCUGCCGCCUCUAACGCUCGGCGAUGACGGUGAUAAUUUCUUUGCCGUAAAUGCCUU ---..((((((((.---......)))).)))).....(((--------------((((((-..((.............))..)))))))))((..(........)..))........... ( -27.82, z-score = -2.62, R) >droWil1.scaffold_181108 1700013 99 - 4707319 ---CCUCAGCACGU---UCGUCCUGAGCCUGAUUUCCGUU--------------AUUGCG-CUGCUGCCGCCGUUGACUCUCGGCGAUGACGGCGACAACUUCUUUGCCGUUAAUGCCUU ---.(((((.((..---..)).)))))..........(((--------------(..(((-(....).)))...))))....((((.(((((((((........))))))))).)))).. ( -31.20, z-score = -2.03, R) >droVir3.scaffold_12822 2292258 117 - 4096053 ---CCUCGGUACCAGCAAUGUCCUGUGCCUGAUUUCCGUUGUUGCGCUGGCAGCGCUGCUGCCGCUGCCGCCGCUGGCGCUGGCUGACGACGGUGACAAUUUCUUUGCCGUUAAUAGCUU ---...(((..(((((........((((..(((.......))))))).((((((((....).)))))))...)))))..)))((((..(((((..(........)..)))))..)))).. ( -44.90, z-score = -1.30, R) >droMoj3.scaffold_6540 26410890 114 - 34148556 --UCAC-AGCGCAAACAAUGUCCUGCGCCUGAUUUCCGUU---UUGGCGCUGCUGUUGCUGCCGCCAUUGCCGCUGGCGCUGGCCGAUGACGGUGACAAUUUCUUUGCCGUUAAUAGCUU --..((-((((((.....)))...(((((.((.......)---).))))).))))).((((..((((.((((...)))).))))...((((((..(........)..)))))).)))).. ( -38.40, z-score = -0.70, R) >droGri2.scaffold_14624 3211294 120 - 4233967 CCACACAAACACAAACAGCGUCCUAUGCCUGAUUUCCGUUGCCGUUGCGGCAUUGCUGCUGCCCCUGCCGCCGUUGGCUUUGGCCGAUGACGGUGACAAUUUCUUUGCCGUUAAUAGCUU .......(((.(((((((.((.....)))))......((..((((.((((((..((....))...))))))(((((((....)))))))))))..))......))))..)))........ ( -36.90, z-score = -1.24, R) >consensus ___CCUCAACGCCC___UGGUCCUGUGCCUGAUUUCCGUU______________AUUGCC_CUGCUGCCGCCGCUGACGCUCGGCGAUGACGGCGAUAAUUUCUUUGCCGUGAAUGCCUU ..........(((.....))).............................................................((((.(.(((((((........))))))).).)))).. (-14.60 = -13.19 + -1.41)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:41 2011