
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,003,963 – 25,004,069 |
| Length | 106 |
| Max. P | 0.699193 |

| Location | 25,003,963 – 25,004,069 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 75.50 |
| Shannon entropy | 0.42787 |
| G+C content | 0.46786 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -15.59 |
| Energy contribution | -16.62 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.607612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
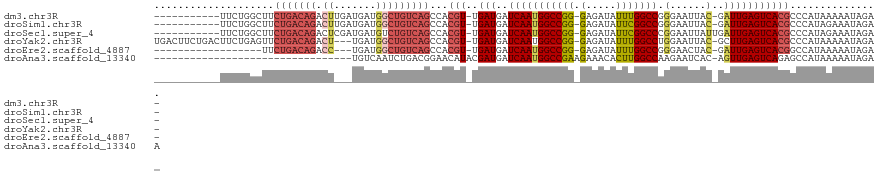
>dm3.chr3R 25003963 106 + 27905053 -----------UUCUGGCUUCUGACAGACUUGAUGAUGGCUGUCAGCCACGU-UGAUGAUCAAUGGCCGG-GAGAUAUUUGGCCGGGAAUUAC-GAUUGAGUCACGCCCAUAAAAAUAGA- -----------(((((((..(((((((.((.......)))))))))...(((-((.....))))))))))-))..(((((....(((....((-......))....)))....)))))..- ( -30.70, z-score = -1.09, R) >droSim1.chr3R 24676257 106 + 27517382 -----------UUCUGGCUUCUGACAGACUUGAUGAUGGCUGUCAGCCACGU-UGAUGAUCAAUGGCCGG-GAGAUAUUCGGCCGGGAAUUAC-GAUUGAGUCACGCCCAUAGAAAUAGA- -----------(((((((..(((((((.((.......)))))))))...(((-((.....))))))))((-(....(((((..((.......)-)..)))))....)))..)))).....- ( -31.70, z-score = -0.91, R) >droSec1.super_4 3868488 107 + 6179234 -----------UUCUGGCUUCUGACAGACUCGAUGAUGUCUGUCAGCCACGU-UGAUGAUCAAUGGCCGG-GAGAUAUUCGGCCCGGAAUUAUUGAUUGAGUCACGCCCAUAGAAAUAGA- -----------(((((((..(((((((((........)))))))))....((-.(((((((((((.((((-(.(.....)..)))))...))))))))..))))))))...)))).....- ( -38.60, z-score = -2.92, R) >droYak2.chr3R 7338118 114 - 28832112 UGACUUCUGACUUCUGAGUUCUGACAGACU---UGAUGGCUGUCAGCCACGU-UGAUGAUCAAUGGCCGG-GAGAUAUUUGGCCUGGAAUUAC-GCUUGAGUCACGCCCAUAAAAAUAGA- .......((((((..((((.(((((((.((---....)))))))))(((.((-((.....))))((((((-(.....))))))))))......-))))))))))................- ( -29.40, z-score = -0.48, R) >droEre2.scaffold_4887 1026 96 - 1626 ------------------UUCUGACAGACC---UGAUGGCUGUCAGCCACGU-UGAUGAUCAAUGGCCGG-GAGAUAUUUGGCCGGGAACUAC-GAUUGAGUCACGGCCAUAAAAAUAGA- ------------------..(((((((.((---....)))))))))((.(((-((.....)))))....)-).......((((((.((.((..-.....)))).))))))..........- ( -30.60, z-score = -2.04, R) >droAna3.scaffold_13340 9459701 87 - 23697760 ---------------------------------UGUCAAUCUGACGGAACAUACGAUGAUCAAUGGCCGAAGAAACACUUGGCCAAGAAUCAC-AGUUGAGUCAGAGCCAUAAAAAUAGAA ---------------------------------......((((((....((.((..((((...(((((((........)))))))...)))).-.)))).))))))............... ( -22.10, z-score = -2.74, R) >consensus ___________UUCUGGCUUCUGACAGACU___UGAUGGCUGUCAGCCACGU_UGAUGAUCAAUGGCCGG_GAGAUAUUUGGCCGGGAAUUAC_GAUUGAGUCACGCCCAUAAAAAUAGA_ ....................(((((((.(........).)))))))..........((((((((((((((........))))))...........))))).)))................. (-15.59 = -16.62 + 1.03)

| Location | 25,003,963 – 25,004,069 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 75.50 |
| Shannon entropy | 0.42787 |
| G+C content | 0.46786 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -15.59 |
| Energy contribution | -17.45 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.699193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
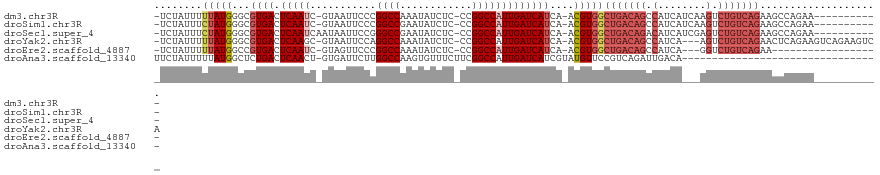
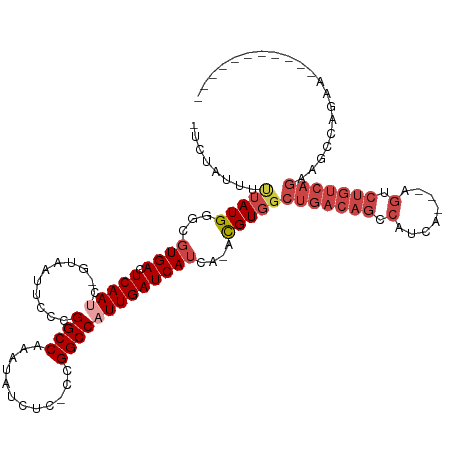
>dm3.chr3R 25003963 106 - 27905053 -UCUAUUUUUAUGGGCGUGACUCAAUC-GUAAUUCCCGGCCAAAUAUCUC-CCGGCCAUUGAUCAUCA-ACGUGGCUGACAGCCAUCAUCAAGUCUGUCAGAAGCCAGAA----------- -(((........(((.((.((......-)).)).)))((((.........-..))))...........-..((..(((((((.(........).)))))))..)).))).----------- ( -24.70, z-score = -0.47, R) >droSim1.chr3R 24676257 106 - 27517382 -UCUAUUUCUAUGGGCGUGACUCAAUC-GUAAUUCCCGGCCGAAUAUCUC-CCGGCCAUUGAUCAUCA-ACGUGGCUGACAGCCAUCAUCAAGUCUGUCAGAAGCCAGAA----------- -.....((((..(((.((.((......-)).)).)))(((((........-.)))))...........-..((..(((((((.(........).)))))))..)).))))----------- ( -27.60, z-score = -0.92, R) >droSec1.super_4 3868488 107 - 6179234 -UCUAUUUCUAUGGGCGUGACUCAAUCAAUAAUUCCGGGCCGAAUAUCUC-CCGGCCAUUGAUCAUCA-ACGUGGCUGACAGACAUCAUCGAGUCUGUCAGAAGCCAGAA----------- -..........(((((((......((((((....(((((..(.....).)-))))..)))))).....-))))..(((((((((........)))))))))...)))...----------- ( -32.50, z-score = -1.89, R) >droYak2.chr3R 7338118 114 + 28832112 -UCUAUUUUUAUGGGCGUGACUCAAGC-GUAAUUCCAGGCCAAAUAUCUC-CCGGCCAUUGAUCAUCA-ACGUGGCUGACAGCCAUCA---AGUCUGUCAGAACUCAGAAGUCAGAAGUCA -............(((.((((((..((-((.......((((.........-..))))..((.....))-))))..(((((((.(....---.).)))))))......).)))))...))). ( -27.30, z-score = -0.56, R) >droEre2.scaffold_4887 1026 96 + 1626 -UCUAUUUUUAUGGCCGUGACUCAAUC-GUAGUUCCCGGCCAAAUAUCUC-CCGGCCAUUGAUCAUCA-ACGUGGCUGACAGCCAUCA---GGUCUGUCAGAA------------------ -..........((((((.(((((....-).))))..))))))....(((.-..((((((((.....))-..))))))(((((((....---)).)))))))).------------------ ( -29.50, z-score = -2.63, R) >droAna3.scaffold_13340 9459701 87 + 23697760 UUCUAUUUUUAUGGCUCUGACUCAACU-GUGAUUCUUGGCCAAGUGUUUCUUCGGCCAUUGAUCAUCGUAUGUUCCGUCAGAUUGACA--------------------------------- ...............((((((.((((.-((((((..((((((((.....))).)))))..)))))).)).))....))))))......--------------------------------- ( -25.30, z-score = -3.17, R) >consensus _UCUAUUUUUAUGGGCGUGACUCAAUC_GUAAUUCCCGGCCAAAUAUCUC_CCGGCCAUUGAUCAUCA_ACGUGGCUGACAGCCAUCA___AGUCUGUCAGAAGCCAGAA___________ ........(((((...((((.(((((...........((((............)))))))))))))....)))))(((((((.(........).))))))).................... (-15.59 = -17.45 + 1.86)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:41 2011