| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,998,518 – 24,998,608 |
| Length | 90 |
| Max. P | 0.990163 |

| Location | 24,998,518 – 24,998,608 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.38 |
| Shannon entropy | 0.47878 |
| G+C content | 0.35345 |
| Mean single sequence MFE | -15.75 |
| Consensus MFE | -10.40 |
| Energy contribution | -10.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.990163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
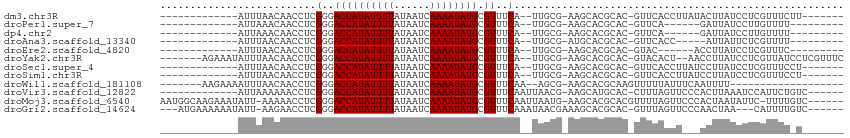
>dm3.chr3R 24998518 90 - 27905053 -------------AUUUAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCA--UUGCG-AAGCACGCAC-GUUCACCUUAUACUUAUCCUCGUUUCUU------- -------------....(((......(..((((((((((......)))))))).))..).--.((((-.....)))).-))).........................------- ( -14.30, z-score = -2.05, R) >droPer1.super_7 2004281 82 - 4445127 -------------AUUAAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCA--UUGCG-AAGCACGCAC-GUUCA------GAUUAUCCUUGUUUU--------- -------------...((((((....(..((((((((((......)))))))).))..).--.((((-.....)))).-.....------........)))))).--------- ( -16.30, z-score = -2.18, R) >dp4.chr2 1840760 82 - 30794189 -------------AUUAAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCA--UUGCG-AAGCACGCAC-GUUCA------GAUUAUCCUUGUUUU--------- -------------...((((((....(..((((((((((......)))))))).))..).--.((((-.....)))).-.....------........)))))).--------- ( -16.30, z-score = -2.18, R) >droAna3.scaffold_13340 9454208 83 + 23697760 -------------AUUUAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCA--UUGCG-AUGCACGCAC-GUUCACC-----AUUAUUCUCGUUUU--------- -------------....(((......(..((((((((((......)))))))).))..).--.((((-.....)))).-)))....-----..............--------- ( -14.30, z-score = -1.67, R) >droEre2.scaffold_4820 7438810 82 + 10470090 -------------AUUUAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCA--UUGCG-AAGCACGCAC-GUAC------ACCUUAUCCUCGUUUC--------- -------------....(((......(..((((((((((......)))))))).))..).--.((((-.....)))).-....------...........)))..--------- ( -14.30, z-score = -1.87, R) >droYak2.chr3R 7332410 101 + 28832112 -------AGAAAUAUUUAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCA--UUGCG-AAGCACGCAC-GUACACU--AACCUUAUCCUCGUUAUCCUCGUUUC -------.(((((...((((......(..((((((((((......)))))))).))..).--.((((-.....)))).-.......--............)))).....))))) ( -17.60, z-score = -2.06, R) >droSec1.super_4 3862967 90 - 6179234 -------------AUUUAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCA--UUGCG-AAGCACGCAC-GUUCACCUUAUCCUUAUCCUCGUUUCCU------- -------------....(((......(..((((((((((......)))))))).))..).--.((((-.....)))).-))).........................------- ( -14.30, z-score = -2.06, R) >droSim1.chr3R 24670067 90 - 27517382 -------------AUUUAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCA--UUGCG-AAGCACGCAC-GUUCACCUUAUCCUUAUCCUCGUUUCCU------- -------------....(((......(..((((((((((......)))))))).))..).--.((((-.....)))).-))).........................------- ( -14.30, z-score = -2.06, R) >droWil1.scaffold_181108 1689211 85 - 4707319 -------AAGAAAAUUUAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCAA--AGCG-AAGCACGCAAGUUUUUAUUUCAAUUUU------------------- -------..((((....(((......(..((((((((((......)))))))).))..)..--.(((-.....)))..)))....))))......------------------- ( -17.10, z-score = -3.10, R) >droVir3.scaffold_12822 2266180 93 - 4096053 -------------AUUAAAAAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCAAUUAACG-AAGCAUGCAC-CUUUAGUUCCCACUUAAAUCCAUUCUGUC------ -------------.............((((.((((((((......)))))))).(((((.......)-))))......-.......))))..................------ ( -14.50, z-score = -2.19, R) >droMoj3.scaffold_6540 26400690 105 - 34148556 AAUGGCAAGAAAUAUU-AAAAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCAAUUAAUG-AAGCACGCACGUUUUAGUUCCCACUAAUAUUC-UUUUGUC------ ...(((((((((((((-(........(..((((((((((......)))))))).))..)......((-((((......))))))........))))))).-)))))))------ ( -21.80, z-score = -2.84, R) >droGri2.scaffold_14624 3200817 100 - 4233967 ---AUGAAAAAAUAUU-AAGAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCAAAUAACGAAAGCACGCAC-GUUUAGUUCCCAACUAA---CAUUUUGUC------ ---.............-..((((...(..((((((((((......)))))))).))..).........((((......-)))).))))........---.........------ ( -13.90, z-score = -0.72, R) >consensus _____________AUUUAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCA__UUGCG_AAGCACGCAC_GUUCACUU___ACUUAUCCUCGUUUU_U_______ ..........................(..((((((((((......)))))))).))..)....................................................... (-10.40 = -10.40 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:39 2011