| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,998,157 – 24,998,264 |
| Length | 107 |
| Max. P | 0.999978 |

| Location | 24,998,157 – 24,998,264 |
|---|---|
| Length | 107 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 62.11 |
| Shannon entropy | 0.78030 |
| G+C content | 0.55882 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -10.52 |
| Energy contribution | -9.79 |
| Covariance contribution | -0.73 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.977919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
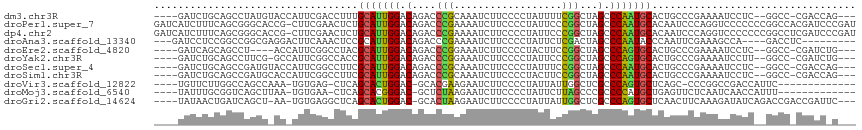
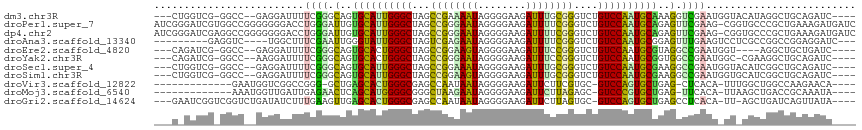
>dm3.chr3R 24998157 107 + 27905053 ----GAUCUGCAGCCUAUGUACCAUUCGACCUUUGCAUUGGACAGACCCGCAAAUCUUCCCCUAUUUUCGGCUAGCCCAAUGCACUGCCCGAAAAUCCUC--GGCC-CGACCAG--- ----....((((.....))))....(((.....((((((((.(((..(((.((((........)))).))))).).))))))))....((((......))--))..-)))....--- ( -21.20, z-score = -0.86, R) >droPer1.super_7 2003855 116 + 4445127 GAUCAUCUUUCAGCGGGCACCG-CUUCGAACUCUGCAUUGGACAGACCCGAAAAUCUUCCCCUAUUCCCGGCUAGCCCAAUGCACAAUCCCAGGUCCCCCCCGGCCACGAUCCCGAU ((((....((((((((...)))-))..)))...((((((((.(((..(((..(((........)))..))))).).))))))))........((((......))))..))))..... ( -27.10, z-score = -0.31, R) >dp4.chr2 1840333 116 + 30794189 GAUCAUCUUUCAGCGGGCACCG-CUUCGAACUCUGCAUUGGACAGACCCGAAAAUCUUCCCCUAUUCCCGGCUAGCCCAAUGCACAAUCCCAGGUCCCCCCCGGCCUCGAUCCCGAU ((((....((((((((...)))-))..)))...((((((((.(((..(((..(((........)))..))))).).)))))))).......(((((......))))).))))..... ( -27.20, z-score = -0.20, R) >droAna3.scaffold_13340 9453822 101 - 23697760 ---GAUCCUCCGGCCGGCGAGGACUUCAAACUCCGCAUUGGACAGACCCGAAAAUCUUCCCCUAUUCUCGACUAGCCCAAUACCCAAUUCGAAAGCCA----GACCUC--------- ---.....((.(((...((((((........)))..(((((.(((...(((.(((........))).))).)).).))))).......)))...))).----))....--------- ( -15.50, z-score = 1.12, R) >droEre2.scaffold_4820 7438451 103 - 10470090 ----GAUCAGCAGCCU----ACCAUUCGGCCUACGCAUUGGACAGACCCGGAAAUCUUCCCCUACUUCCGGCUAGCCCAGUGCACUGCCCGAAAAUCCUC--GGCC-CGAUCUG--- ----((((.((((((.----.......)))....(((((((.(((..((((((............)))))))).).)))))))..)))((((......))--))..-.))))..--- ( -32.40, z-score = -3.04, R) >droYak2.chr3R 7332031 106 - 28832112 ----GAUCUGCAGCCUUCG-GCCAUUCGGCCACCGCAUUGGACAGACCCGGAAAUCUUCCCCUAUUCCCGGCUAGCCCAGUGCACUGCCCGAAAAUCCUU--GGCC-CGAUCUG--- ----((((.((((.....(-(((....))))...(((((((.(((.((.((((...........)))).)))).).))))))).))))((((......))--))..-.))))..--- ( -36.10, z-score = -2.92, R) >droSec1.super_4 3862640 107 + 6179234 ----GAUCUGCAGCCGAUGUACCAUUCGGCCUUCGCAUUGGACAGACCCGCAAAUCUUCCCCUAUUUCCGGCUAGCCCAAUGCACUGCCCGAAAAUCCUC--GGCC-CGACCAG--- ----.....((((((((........)))))....(((((((.(((..(((.((((........)))).))))).).)))))))..)))((((......))--))..-.......--- ( -28.00, z-score = -1.77, R) >droSim1.chr3R 24669728 107 + 27517382 ----GAUCUGCAGCCGAUGCACCAUUCGGCCUUCGCAUUGGACAGACCCGCAAAUCUUCCCCUACUUCCGGCUAGCCCAAUGCACUGCCCGAAAAUCCUC--GGCC-CGACCAG--- ----.....((((((((........)))))....(((((((.(((.((.....................)))).).)))))))..)))((((......))--))..-.......--- ( -25.40, z-score = -0.81, R) >droVir3.scaffold_12822 2265766 96 + 4096053 ----UGUUCUUGGCCAGCCAAA-UGUGAG-CUCAGCACUGGAC-GCACGAAGAAUCUUCCCCUAUUAUUGGCUCGCCCAGUGCUCAGC-CCCGGCCGACCAUUC------------- ----.....((((((.((....-.))..(-((.((((((((..-((..((((...)))).((.......))...)))))))))).)))-...))))))......------------- ( -29.40, z-score = -1.38, R) >droMoj3.scaffold_6540 26400197 97 + 34148556 ----UAUUUGCGGUCAGCUUAA-UGUGAA-CUCAGCACGGGAC-GCUCUAAGAAUCUUCCCCUAUUCUUAGCCCGCCCCAUGCUGAGUUCUCAAUCAACCAUUU------------- ----.....((.....))....-((.(((-(((((((.(((.(-(..((((((((........))))))))..)).))).)))))))))).))...........------------- ( -35.60, z-score = -6.25, R) >droGri2.scaffold_14624 3200422 107 + 4233967 ----UAUAACUGAUCAGCU-AA-UGUGAGGCUCAGCACUGGAC-GCACUAAGAAUCUUCCCCUAUUAUUGGCUCGCCCAGUGCUCAACUUCAAAGAUAUCAGACCGACCGAUUC--- ----.....(((((...((-..-..(((((...((((((((.(-(..((((.(((........))).))))..)).))))))))...))))).))..)))))............--- ( -27.40, z-score = -2.00, R) >consensus ____GAUCUGCAGCCGGCG_AC_AUUCGGCCUCCGCAUUGGACAGACCCGAAAAUCUUCCCCUAUUCCCGGCUAGCCCAAUGCACAGCCCGAAAACCCUC__GGCC_CGAUC_G___ ..................................(((((((.(....(((..................)))...).))))))).................................. (-10.52 = -9.79 + -0.73)
| Location | 24,998,157 – 24,998,264 |
|---|---|
| Length | 107 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 62.11 |
| Shannon entropy | 0.78030 |
| G+C content | 0.55882 |
| Mean single sequence MFE | -44.88 |
| Consensus MFE | -26.00 |
| Energy contribution | -24.35 |
| Covariance contribution | -1.65 |
| Combinations/Pair | 1.87 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.56 |
| SVM RNA-class probability | 0.999978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
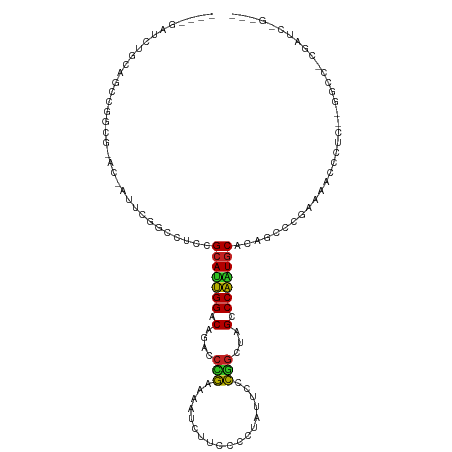
>dm3.chr3R 24998157 107 - 27905053 ---CUGGUCG-GGCC--GAGGAUUUUCGGGCAGUGCAUUGGGCUAGCCGAAAAUAGGGGAAGAUUUGCGGGUCUGUCCAAUGCAAAGGUCGAAUGGUACAUAGGCUGCAGAUC---- ---..((((.-((((--...(((((((((.(..((((((((((.(.(((.((((........)))).))).)..))))))))))..).))))).))).)...))))...))))---- ( -37.10, z-score = -1.93, R) >droPer1.super_7 2003855 116 - 4445127 AUCGGGAUCGUGGCCGGGGGGGACCUGGGAUUGUGCAUUGGGCUAGCCGGGAAUAGGGGAAGAUUUUCGGGUCUGUCCAAUGCAGAGUUCGAAG-CGGUGCCCGCUGAAAGAUGAUC .....((((((...(((.(((.((((.(((((.((((((((((.(.(((..(((........)))..))).)..)))))))))).))))).)..-.))).))).)))....)))))) ( -50.20, z-score = -3.20, R) >dp4.chr2 1840333 116 - 30794189 AUCGGGAUCGAGGCCGGGGGGGACCUGGGAUUGUGCAUUGGGCUAGCCGGGAAUAGGGGAAGAUUUUCGGGUCUGUCCAAUGCAGAGUUCGAAG-CGGUGCCCGCUGAAAGAUGAUC .....(((((....(((.(((.((((.(((((.((((((((((.(.(((..(((........)))..))).)..)))))))))).))))).)..-.))).))).))).....))))) ( -48.60, z-score = -2.76, R) >droAna3.scaffold_13340 9453822 101 + 23697760 ---------GAGGUC----UGGCUUUCGAAUUGGGUAUUGGGCUAGUCGAGAAUAGGGGAAGAUUUUCGGGUCUGUCCAAUGCGGAGUUUGAAGUCCUCGCCGGCCGGAGGAUC--- ---------..((.(----(((((((((((((..(((((((((.(.((((((((........)))))))).)..)))))))))..))))))))).....)))))))........--- ( -37.30, z-score = -1.58, R) >droEre2.scaffold_4820 7438451 103 + 10470090 ---CAGAUCG-GGCC--GAGGAUUUUCGGGCAGUGCACUGGGCUAGCCGGAAGUAGGGGAAGAUUUCCGGGUCUGUCCAAUGCGUAGGCCGAAUGGU----AGGCUGCUGAUC---- ---..(((((-((((--.......(((((.(..((((.(((((.(.((((((((........)))))))).)..))))).))))..).)))))....----.)))..))))))---- ( -44.20, z-score = -2.81, R) >droYak2.chr3R 7332031 106 + 28832112 ---CAGAUCG-GGCC--AAGGAUUUUCGGGCAGUGCACUGGGCUAGCCGGGAAUAGGGGAAGAUUUCCGGGUCUGUCCAAUGCGGUGGCCGAAUGGC-CGAAGGCUGCAGAUC---- ---..((((.-((((--..((...(((((.((.((((.(((((.(.((((((((........)))))))).)..))))).)))).)).)))))...)-)...))))...))))---- ( -49.70, z-score = -3.69, R) >droSec1.super_4 3862640 107 - 6179234 ---CUGGUCG-GGCC--GAGGAUUUUCGGGCAGUGCAUUGGGCUAGCCGGAAAUAGGGGAAGAUUUGCGGGUCUGUCCAAUGCGAAGGCCGAAUGGUACAUCGGCUGCAGAUC---- ---..((((.-((((--((..((((((((.(..((((((((((.(.(((.((((........)))).))).)..))))))))))..).))))).)))...))))))...))))---- ( -45.90, z-score = -3.44, R) >droSim1.chr3R 24669728 107 - 27517382 ---CUGGUCG-GGCC--GAGGAUUUUCGGGCAGUGCAUUGGGCUAGCCGGAAGUAGGGGAAGAUUUGCGGGUCUGUCCAAUGCGAAGGCCGAAUGGUGCAUCGGCUGCAGAUC---- ---..((((.-((((--(((.((((((((.(..((((((((((.(.(((.((((........)))).))).)..))))))))))..).))))).))).).))))))...))))---- ( -46.70, z-score = -3.16, R) >droVir3.scaffold_12822 2265766 96 - 4096053 -------------GAAUGGUCGGCCGGG-GCUGAGCACUGGGCGAGCCAAUAAUAGGGGAAGAUUCUUCGUGC-GUCCAGUGCUGAG-CUCACA-UUUGGCUGGCCAAGAACA---- -------------...((((((((((((-(((.(((((((((((..((........))((((...))))...)-)))))))))).))-)))...-..))))))))))......---- ( -49.00, z-score = -5.05, R) >droMoj3.scaffold_6540 26400197 97 - 34148556 -------------AAAUGGUUGAUUGAGAACUCAGCAUGGGGCGGGCUAAGAAUAGGGGAAGAUUCUUAGAGC-GUCCCGUGCUGAG-UUCACA-UUAAGCUGACCGCAAAUA---- -------------....((((...((.(((((((((((((((((..((((((((........))))))))..)-)))))))))))))-))).))-...))))...........---- ( -47.20, z-score = -7.24, R) >droGri2.scaffold_14624 3200422 107 - 4233967 ---GAAUCGGUCGGUCUGAUAUCUUUGAAGUUGAGCACUGGGCGAGCCAAUAAUAGGGGAAGAUUCUUAGUGC-GUCCAGUGCUGAGCCUCACA-UU-AGCUGAUCAGUUAUA---- ---.....((((((.(((((.....(((.(((.(((((((((((.((......((((((....)))))))).)-)))))))))).))).))).)-))-)))))))).......---- ( -37.80, z-score = -2.57, R) >consensus ___C_GAUCG_GGCC__GAGGAUUUUCGGGCUGUGCAUUGGGCUAGCCGGGAAUAGGGGAAGAUUUUCGGGUCUGUCCAAUGCGGAGGCCGAAU_GU_CGCCGGCUGCAGAUC____ .........................((((.(..((((((((((...((((((((........))))))))....))))))))))..).))))......................... (-26.00 = -24.35 + -1.65)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:38 2011