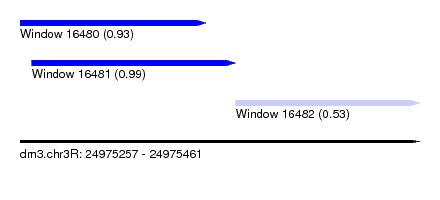
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,975,257 – 24,975,461 |
| Length | 204 |
| Max. P | 0.992383 |

| Location | 24,975,257 – 24,975,352 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 69.10 |
| Shannon entropy | 0.65960 |
| G+C content | 0.48076 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -12.32 |
| Energy contribution | -12.10 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.47 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
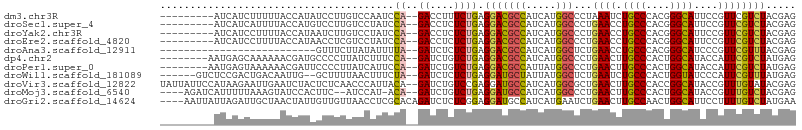
>dm3.chr3R 24975257 95 + 27905053 ---------AUCAUCUUUUUACCAUAUCCUUGUCCAAUCCA--GACCUUUCUGAGGACGCCAUCAUGGCCCUAAAUCUGCCCACGGGCAUUCCGUUCGUCUACGAG ---------...................(((((......((--((....)))).(((((((.....)))........((((....))))........))))))))) ( -20.00, z-score = -1.43, R) >droSec1.super_4 3839825 95 + 6179234 ---------AUCAUCAUUUUACCAUGUCCUUGUCCUAUCCA--GACCUCUCUGAGGACGCCAUCAUGGCCCUGAACCUGCCCACGGGCAUUCCGUUCGUCUACGAG ---------....((......(((((....((((((...((--((....))))))))))....)))))....((((.((((....))))....))))......)). ( -25.80, z-score = -2.30, R) >droYak2.chr3R 7309070 95 - 28832112 ---------AUCAUCCUUUUACCAUAAUCUUGUCCUAUCCA--GACCUCUCUGAGGACGCCAUCAUGGCCCUGAACCUGCCCACGGGCAUUCCGUUCGUCUACGAG ---------.((.........((((.((..((((((...((--((....))))))))))..)).))))....((((.((((....))))....))))......)). ( -22.90, z-score = -1.87, R) >droEre2.scaffold_4820 7413623 95 - 10470090 ---------AUCAUCCUUUUACCAUAACCUCGUCCUAUCCA--GACCUCUCUGAGGACGCCAUCAUGGCCCUGAACCUGCCCACGGGCAUUCCGUUCGUCUACGAG ---------...................(((((......((--((....)))).(((((((.....)))...((((.((((....))))....))))))))))))) ( -25.10, z-score = -2.67, R) >droAna3.scaffold_12911 4034207 78 + 5364042 --------------------------GUUUCUUAUAUUUUA--GAUCUCUCUGAGGACGCCAUCAUGGCUCUGAACCUGCCCACGGGCAUCCCGUUCGUUUACGAG --------------------------............(((--((....)))))(((((((.....)))...((((.((((....))))....))))))))..... ( -17.60, z-score = -0.68, R) >dp4.chr2 8719590 96 + 30794189 --------AAUGAGCAAAAAACGAUGCCCCUUAUCUUUCCA--GAUCUGUCUGAGGACGCCAUCAUGGCCCUGAACUUGCCCACUGGCAUACCAUUCGUCUAUGAG --------.....(((........)))..(((((.....((--((....)))).(((((((.....)))...(((..((((....)))).....)))))))))))) ( -18.40, z-score = -0.04, R) >droPer1.super_0 2533135 96 + 11822988 --------AAUGAGUAAAAAACGAUUCCCCUUAUCAUUCCA--GAUCUGUCUGAGGACGCCAUUAUGGCCCUGAACUUGCCCACUGGCAUACCAUUCGUCUAUGAG --------...((((........))))......((((..((--((....)))).(((((((.....)))...(((..((((....)))).....))))))))))). ( -17.80, z-score = -0.26, R) >droWil1.scaffold_181089 5499211 96 + 12369635 ------GUCUCCGACUGACAAUUG--GCUUUUAACUUUCUA--GAUCUCUCUGAGGAUGCUAUUAUGGCUCUGAAUCUGCCCACUGGUAUCCCAUUCGUUUAUGAG ------(((.......))).....--.(((..(((....((--((....)))).((((((((....(((.........)))...)))))))).....)))...))) ( -17.60, z-score = 0.18, R) >droVir3.scaffold_12822 2240856 104 + 4096053 UAUUAUUCCAUAAGAAUUGAAUCUACUCUCAACCCAUUACA--GAUCUGUCCGAGGAUGCCAUCAUGGCGCUGAACUUGCCCACCGGCAUACCGUUUGUAUACGAG ................((((........)))).....((((--((((.....))(((((((....(((.((.......)))))..))))).)).))))))...... ( -17.30, z-score = 0.42, R) >droMoj3.scaffold_6540 23194077 97 - 34148556 ----AGAUCAUUUUUAAAGUAUCCACUUC--AUCCAU-ACA--GAUCUGUCUGAGGAUGCCAUCAUGGCCCUGAACUUGCCCACUGGCAUACCGUUUGUCUACGAG ----..............(((..((....--......-.((--((....)))).((((((((....(((.........)))...)))))).))...))..)))... ( -19.20, z-score = -0.11, R) >droGri2.scaffold_14624 3177715 102 + 4233967 ----AAUUAUUAGAUUGCUAACUAUUGUUGUUAACCUCGCACAGAUCUCUCGGAGGAUGCCAUCAUGAAUCUGAACUUGCCAACUGGCAUUCCUUUUGUCUAUGAA ----.....(((((((..((((.......)))).((((.(..((....)).)))))...........)))))))...((((....))))................. ( -16.10, z-score = 0.80, R) >consensus _________AUCAUCAUUUAACCAUAUCCUUUUCCAUUCCA__GAUCUCUCUGAGGACGCCAUCAUGGCCCUGAACCUGCCCACGGGCAUUCCGUUCGUCUACGAG ......................................................(((((((.....)))...((((.((((....))))....))))))))..... (-12.32 = -12.10 + -0.22)
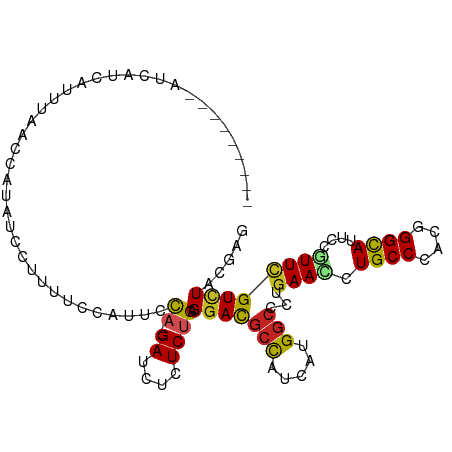
| Location | 24,975,263 – 24,975,367 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 72.81 |
| Shannon entropy | 0.60934 |
| G+C content | 0.49588 |
| Mean single sequence MFE | -26.53 |
| Consensus MFE | -16.24 |
| Energy contribution | -16.25 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.992383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24975263 104 + 27905053 -------UUUUUACCAUAUCCUUGUCCAAU-CCAGACCUUUCUGAGGACGCCAUCAUGGCCCUAAAUCUGCCCACGGGCAUUCCGUUCGUCUACGAGCUGGACGAGAACUUC -------.............((((((((..-.((((....)))).(((.(((.....)))........((((....)))).)))(((((....)))))))))))))...... ( -33.40, z-score = -3.63, R) >droSim1.chr3R 13328918 79 + 27517382 ---------------------------------AUGCAUCUCUGACAAGGACAUAAUGAGCCUCAACCUGCCCACGGGCAUUCCUUUCGUCUACGAACUGGACGAGAACCUC ---------------------------------.....((((.((.(((((.....((.....))...((((....)))).)))))))(((((.....)))))))))..... ( -19.70, z-score = -1.58, R) >droSec1.super_4 3839831 104 + 6179234 -------AUUUUACCAUGUCCUUGUCCUAU-CCAGACCUCUCUGAGGACGCCAUCAUGGCCCUGAACCUGCCCACGGGCAUUCCGUUCGUCUACGAGCUGGACGAGAACUUC -------..........((.(((((((...-.(((.((((...))))..(((.....))).)))....((((....))))....(((((....))))).))))))).))... ( -33.30, z-score = -2.64, R) >droYak2.chr3R 7309076 104 - 28832112 -------CUUUUACCAUAAUCUUGUCCUAU-CCAGACCUCUCUGAGGACGCCAUCAUGGCCCUGAACCUGCCCACGGGCAUUCCGUUCGUCUACGAGCUGGACGAGAACUUC -------............((((((((...-.(((.((((...))))..(((.....))).)))....((((....))))....(((((....))))).))))))))..... ( -34.00, z-score = -3.39, R) >droEre2.scaffold_4820 7413629 104 - 10470090 -------CUUUUACCAUAACCUCGUCCUAU-CCAGACCUCUCUGAGGACGCCAUCAUGGCCCUGAACCUGCCCACGGGCAUUCCGUUCGUCUACGAGCUGGACGAGAACUUC -------.............(((((((...-.(((.((((...))))..(((.....))).)))....((((....))))....(((((....))))).)))))))...... ( -34.50, z-score = -3.52, R) >droAna3.scaffold_12911 4034208 92 + 5364042 --------------------UUUCUUAUAUUUUAGAUCUCUCUGAGGACGCCAUCAUGGCUCUGAACCUGCCCACGGGCAUCCCGUUCGUUUACGAGCUGGACGAGAACUUC --------------------.(((((....((((((((((...))))..(((.....)))))))))..((((....))))(((.(((((....))))).))).))))).... ( -27.30, z-score = -2.05, R) >dp4.chr2 8719597 104 + 30794189 -------AAAAAACGAUGCCCCUUAUC-UUUCCAGAUCUGUCUGAGGACGCCAUCAUGGCCCUGAACUUGCCCACUGGCAUACCAUUCGUCUAUGAGCUGGACGAGAACUUC -------........(((((.......-....((((....))))(((..(((.....)))))).............)))))....((((((((.....))))))))...... ( -24.20, z-score = -0.92, R) >droPer1.super_0 2533142 104 + 11822988 -------AAAAAACGAUUCCCCUUAUC-AUUCCAGAUCUGUCUGAGGACGCCAUUAUGGCCCUGAACUUGCCCACUGGCAUACCAUUCGUCUAUGAGCUGGACGAGAACUUC -------.......(((.......)))-....((((....)))).((..(((.....)))........((((....))))..)).((((((((.....))))))))...... ( -22.30, z-score = -0.62, R) >droWil1.scaffold_181089 5499217 105 + 12369635 ------GACUGACAAUUGGCUUUUAACUUUC-UAGAUCUCUCUGAGGAUGCUAUUAUGGCUCUGAAUCUGCCCACUGGUAUCCCAUUCGUUUAUGAGUUGGAUGAAAACUUC ------......((.(..((((..(((....-((((....)))).((((((((....(((.........)))...)))))))).....)))...))))..).))........ ( -22.40, z-score = -0.71, R) >droVir3.scaffold_12822 2240863 112 + 4096053 CCAUAAGAAUUGAAUCUACUCUCAACCCAUUACAGAUCUGUCCGAGGAUGCCAUCAUGGCGCUGAACUUGCCCACCGGCAUACCGUUUGUAUACGAGCUGGAUGAGAACUUC ...................(((((.((..................(((((((....(((.((.......)))))..))))).))(((((....))))).)).)))))..... ( -25.80, z-score = -0.69, R) >droMoj3.scaffold_6540 23194085 104 - 34148556 -------UUUAAAGUAUCCACUUCAUCCAU-ACAGAUCUGUCUGAGGAUGCCAUCAUGGCCCUGAACUUGCCCACUGGCAUACCGUUUGUCUACGAGCUGGACGAGAACUUC -------..............(((.((((.-.((((....)))).((((((((....(((.........)))...)))))).))(((((....))))))))).)))...... ( -25.60, z-score = -0.91, R) >droGri2.scaffold_14624 3177726 106 + 4233967 ------UGCUAACUAUUGUUGUUAACCUCGCACAGAUCUCUCGGAGGAUGCCAUCAUGAAUCUGAACUUGCCAACUGGCAUUCCUUUUGUCUAUGAACUCGAUGAAAACUUC ------.......(((((..(((......((((((((.((...((((...)).))..)))))))....((((....)))).......))).....))).)))))........ ( -15.80, z-score = 0.98, R) >consensus _______AUUUAACCAUACCCUUGACCUAU_CCAGAUCUCUCUGAGGACGCCAUCAUGGCCCUGAACCUGCCCACGGGCAUUCCGUUCGUCUACGAGCUGGACGAGAACUUC ................................((((....)))).((..(((.....)))........((((....))))..)).((((((((.....))))))))...... (-16.24 = -16.25 + 0.01)
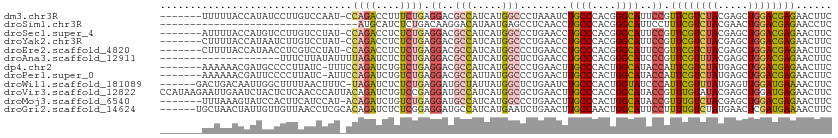
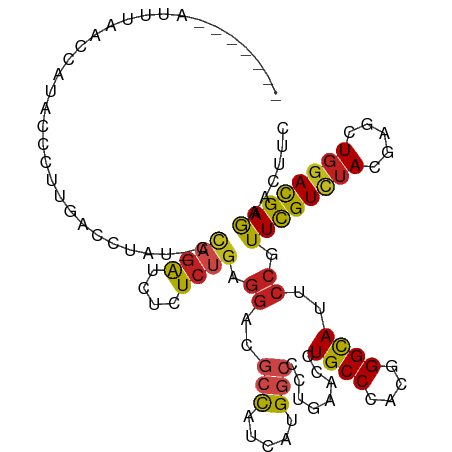
| Location | 24,975,367 – 24,975,461 |
|---|---|
| Length | 94 |
| Sequences | 15 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 80.97 |
| Shannon entropy | 0.42974 |
| G+C content | 0.57771 |
| Mean single sequence MFE | -33.97 |
| Consensus MFE | -19.45 |
| Energy contribution | -19.09 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.73 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.531256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24975367 94 + 27905053 -------AAGCCGGUGGUGUCCAUGCAGUUCCUCGGCGACGAGGAGACCGUGAAGAAGGCCAUCGAGGCUGUGGCCGCCCAGGGCAAGGCCAAGUAAGCAC -------....(((((((.((((((...(((((((....)))))))..))))..))..)))))))..(((.((((((((...)))..)))))....))).. ( -41.40, z-score = -2.36, R) >droSim1.chr3R_random 1212692 94 + 1307089 -------AAGCCGGUGGUGUCGAUGCAGUUCCUCGGCGACGAGGAGACCGUGAAGAAGGCCAUCGAGGCCGUUGCCGCCCAGGGCAAGGCCAAGUAAGCAC -------..(((((((((.((..(((.((((((((....)))))).)).)))..))..))))))).((((.(((((......)))))))))......)).. ( -43.50, z-score = -3.33, R) >droSec1.super_4 3839935 94 + 6179234 -------AAGCCGGUGGUGUCGAUGCAGUUCCUCGGCGACGAGGAGACCGUGAAAAAGGCCAUCGAGGCCGUUGCCGCCCAGGGCAAGGCCAAGUAAGCAC -------..(((((((((.((.(((...(((((((....)))))))..))))).....))))))).((((.(((((......)))))))))......)).. ( -41.20, z-score = -2.73, R) >droYak2.chr3R 7309180 94 - 28832112 -------AAGCCGGUGGUGUCCAUGCAGUUCCUCGGCGACGAGGAGACUGUGAAGAAGGCCAUCGAGGCGGUGGCUGCCCAGGGCAAGGCCAAGUAAGCAC -------.....(((..(((((..(((((((((((....)))))).)))))......((((((((...)))))))).....)))))..))).......... ( -41.90, z-score = -2.51, R) >droEre2.scaffold_4820 7413733 94 - 10470090 -------AAGCCGGUGGUGUCCAUGCAGUUCCUCGGCGACGAGGAGACUGUGAAGAAGGCCAUCGAGGCUGUGGCUGCCCAGGGCAAGGCCAAGUAAGCAG -------....(((((((.((..((((((((((((....)))))).))))))..))..)))))))..(((.((((((((...)))..)))))....))).. ( -42.70, z-score = -2.81, R) >droAna3.scaffold_12911 4034300 94 + 5364042 -------AAGCCCGUGGUGUCGAUGCAGUUCCUCGGCGACGAGGAGACCGUGAAGAAGGCCAUUGAGGCUGUCGCCGCCCAGGGCAAGGCCAAGUAAGCUA -------..((((..((((.((((((.((((((((....)))))).)).))......((((.....)))))))).))))..))))..(((.......))). ( -38.40, z-score = -1.79, R) >dp4.chr2 8719701 92 + 30794189 -------AAGCCAGUGGUGUCCAUGCAGUUCCUCGGCGAUGAGGAGACCGUCAAGAAGGCCAUCGAGGCUGUCGCUGCCCAGGGCAAGGCCAAGUAAAC-- -------..(((.....(((((..(((((((((((....)))))).........((.((((.....)))).)))))))...))))).))).........-- ( -31.30, z-score = -0.61, R) >droPer1.super_0 2533246 92 + 11822988 -------AAGCCAGUGGUGUCCAUGCAGUUCCUCGGCGAUGAGGAGACCGUCAAGAAGGCCAUCGAGGCUGUCGCUGCCCAGGGCAAGGCCAAGUAAAC-- -------..(((.....(((((..(((((((((((....)))))).........((.((((.....)))).)))))))...))))).))).........-- ( -31.30, z-score = -0.61, R) >droWil1.scaffold_181089 5499322 91 + 12369635 -------AAGCCUGUGGUGUCCAUGCAGUUUUUGGGUGAUGAGGAGACAGUCAAGAAGGCCAUUGAAGCCGUGGCUGCUCAGGGCAAAGCCAAGUAAA--- -------..((((....(..(((.........)))..).....(((.((((((....(((.......))).))))))))).)))).............--- ( -25.50, z-score = 0.47, R) >droVir3.scaffold_12822 2240975 93 + 4096053 -------AAGCCCGUUGUAUCCAUGCAGUUCUUGGGUGACGAGGAGACCGUCAAGAAGGCCAUCGAAGCCGUUGCUGCCCAAGGCAAGGCCAAGUAAAUG- -------..(((...((((....))))((.(((((((((((.(....))))).....(((.......)))......)))))))))..)))..........- ( -28.70, z-score = -1.07, R) >droMoj3.scaffold_6540 23194189 93 - 34148556 -------AAGCCCGUGGUAUCCAUGCAGUUCCUUGGCGACGAGGAGACAGUCAAGAAGGCCAUCGAAGCUGUUGCUGCCCAGGGCAAGGCCAAGUAAACG- -------..((((((((...))))(((((((((((....))))))((((((...((......))...)))))))))))...))))...............- ( -32.60, z-score = -1.52, R) >droGri2.scaffold_14624 3177832 94 + 4233967 -------AAGCCAGUUGUCUCCAUGCAGUUCCUCGGCGACGAGGAGACCGUGAAGAAGGCCAUCGAAGCUGUCGCUGCCCAGGGCAAGGCCAAGUAAAUCC -------..(((..((((((....(((((((((((....))))))(((.((...((......))...)).))))))))...)))))))))........... ( -32.30, z-score = -1.28, R) >anoGam1.chr2R 57986429 94 - 62725911 -------AAGCCGGUCGUGUCGAUGAAGUUCCUCGGCGAUGAGGAGACGGUCCGCAAGGCGAUCGAAUCCGUCGCUAACCAGGGCAAGGCCAAGUAAGGCG -------..((((((...(.(((((...(((((((....))))))).((((((....))..))))....))))))..)))..(((...)))......))). ( -32.30, z-score = -0.23, R) >apiMel3.GroupUn 379579703 90 - 399230636 -------AAGCCUGUUGUAUCUAUGAAAUUCUUAGGCGAUGAAGAAACUGUAAAAGCAGCAAUGGCUGCAGUUGCUGCACAAGCAAAAGUUAAAUAA---- -------..(((((..((.((...)).))...)))))........((((......(((((....)))))..(((((.....))))).))))......---- ( -20.90, z-score = -0.37, R) >triCas2.chrUn_11 412319 100 + 1365317 AAACCCACAGCUAAUGGCAGUCUU-CAGUUCUUGGGGGAUCCAGAAACAGUUAAAAAGGCCAUGGAAGCGGUGGCCAACCAAGGCAAAGCCAAGUAAACGC .........(((..((((.(((((-..(((..(((.....)))..))).........((((((.(...).))))))....)))))...)))))))...... ( -25.50, z-score = 0.18, R) >consensus _______AAGCCGGUGGUGUCCAUGCAGUUCCUCGGCGACGAGGAGACCGUGAAGAAGGCCAUCGAGGCUGUCGCUGCCCAGGGCAAGGCCAAGUAAACA_ .........(((.(((((.((...((..(((((((....)))))))...))...))..)))))...(((....)))......)))................ (-19.45 = -19.09 + -0.35)
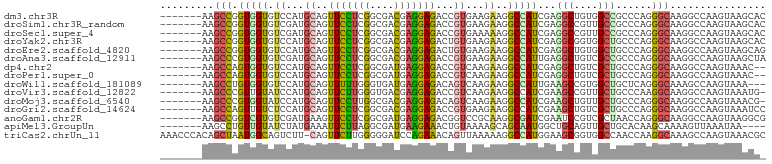

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:36 2011