
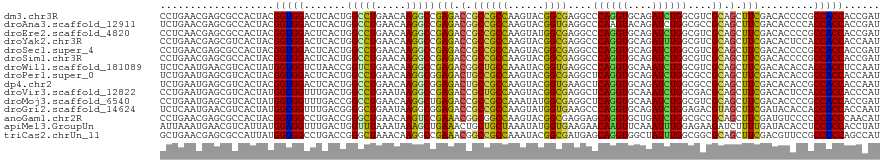
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,974,887 – 24,975,007 |
| Length | 120 |
| Max. P | 0.587759 |

| Location | 24,974,887 – 24,975,007 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.23 |
| Shannon entropy | 0.38587 |
| G+C content | 0.60611 |
| Mean single sequence MFE | -43.72 |
| Consensus MFE | -29.28 |
| Energy contribution | -28.15 |
| Covariance contribution | -1.13 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.587759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
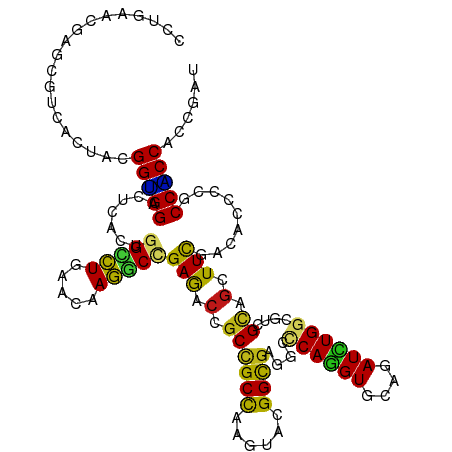
>dm3.chr3R 24974887 120 + 27905053 CCUGAACGAGCGCCACUACGGUGGACUCACUGGCCUGAACAAGGCCGAGACCGCCGCCAAGUACGGCGAGGCCCAGGUGCAGAUCUGGCGUCGCAGCUUCGACACCCCGCCACCACCGAU ..................((((((.(((...(((((.....))))))))..(((((.......))))).(((((((((....)))))).((((......)))).....))).)))))).. ( -45.80, z-score = -1.28, R) >droAna3.scaffold_12911 4033822 120 + 5364042 UCUGAACGAGCGCCACUACGGUGGACUCACUGGCCUGAACAAGGCCGAGACGGCCGCCAAGUACGGUGAGGCCCAAGUACAGAUCUGGCGCCGCAGCUUCGACACCCCACCACCACCGAU .(((..((.((((((.(((..(((.(((((((..((......(((((...)))))....))..)))))))..))).)))......)))))))))))........................ ( -39.30, z-score = -0.47, R) >droEre2.scaffold_4820 7413256 120 - 10470090 CCUCAACGAGCGCCACUACGGUGGACUCACUGGCCUGAACAAGGCCGAGACCGCCGCCAAGUAUGGCGAGGCCCAGGUGCAGAUCUGGCGUCGCAGCUUCGACACCCCGCCACCACCGAU .(((...(((..((((....)))).)))...(((((.....)))))))).....(((((....))))).(((((((((....)))))).((((......)))).....)))......... ( -46.80, z-score = -1.59, R) >droYak2.chr3R 7308701 120 - 28832112 CCUGAACGAGCGUCACUACGGUGGACUCACUGGCCUGAACAAGGCCGAGACCGCCGCCAAGUACGGCGAGGCCCAGGUGCAGAUUUGGCGUCGCAGCUUCGACACUCCACCACCACCAAU .......(((.(((....((((((.(((...(((((.....)))))))).))))))..((((...((((.(((.((((....))))))).)))).)))).))).)))............. ( -45.10, z-score = -1.83, R) >droSec1.super_4 3839458 120 + 6179234 CCUGAACGAGCGCCACUACGGUGGACUCACUGGCCUGAACAAGGCCGAGACCGCCGCCAAGUACGGCGAGGCCCAGGUGCAGAUCUGGCGUCGCAGCUUCGACACCCCGCCACCACCGAU ..................((((((.(((...(((((.....))))))))..(((((.......))))).(((((((((....)))))).((((......)))).....))).)))))).. ( -45.80, z-score = -1.28, R) >droSim1.chr3R 24651354 120 + 27517382 CCUGAACGAGCGCCACUACGGUGGACUCACUGGCCUGAACAAGGCCGAGACCGCCGCCAAGUACGGCGAGGCCCAGGUGCAGAUCUGGCGUCGCAGCUUCGACACCCCGCCACCACCGAU ..................((((((.(((...(((((.....))))))))..(((((.......))))).(((((((((....)))))).((((......)))).....))).)))))).. ( -45.80, z-score = -1.28, R) >droWil1.scaffold_181089 5498835 120 + 12369635 UCUCAAUGAACGUCACUAUGGUGGUCUAACCGGUCUGAACAAGGCCGAGACGGCUGCCAAAUACGGUGAGGCCCAGGUGCAAAUCUGGCGUCGCAGCUUCGACACACCACCACCUCCAAU ..................((((((((....((((((.....)))))).((.((((((((.......)).(((((((((....)))))).)))))))))))))).)))))........... ( -40.20, z-score = -1.39, R) >droPer1.super_0 2532767 120 + 11822988 UCUGAAUGAGCGUCACUACGGUGGACUCACUGGCCUGAACAAGGCGGAGACUGCCGCCAAGUACGGCGAGGCUCAGGUGCAGAUCUGGCGCCGCAGCUUCGACACACCGCCACCACCAAU ......((((..((((....)))).)))).(((((((.((..(((((......)))))..)).)))(((((((..(((((.......)))))..))))))).......))))........ ( -45.10, z-score = -1.08, R) >dp4.chr2 8719222 120 + 30794189 UCUGAAUGAGCGUCACUACGGUGGACUCACUGGCCUGAACAAGGCGGAGACUGCCGCCAAGUACGGUGAAGCUCAGGUGCAGAUCUGGCGCCGCAGCUUCGACACACCGCCACCACCAAU ......((((..((((....)))).)))).(((((((.((..(((((......)))))..)).)))(((((((..(((((.......)))))..))))))).......))))........ ( -43.80, z-score = -1.09, R) >droVir3.scaffold_12822 2240494 120 + 4096053 CCUGAAUGAGCGUCACUAUGGCGGUUUGACUGGCCUGAAUAAGGCCGAGACCGCUGCCAAGUACGGCGAGGCUCAGGUGCAAAUCUGGCGACGCAGCUUCGACACUCCACCGCCACCCAU ......(((((.((.....((((((((...((((((.....))))))))))))))(((......))))).)))))((((....((.(((......)))..)).....))))......... ( -44.80, z-score = -1.57, R) >droMoj3.scaffold_6540 23193711 120 - 34148556 CCUGAAUGAGCGUCACUAUGGUGGUUUGACCGGCCUGAACAAGGCUGAGACCGCCGCCAAAUAUGGCGAGGCUCAGGUGCAAAUCUGGCGUCGCAGCUUCGACACCCCGCCACCACCGAU ......(((((.((.....((((((((...((((((.....))))))))))))))((((....)))))).)))))((((.......((.((((......)))).)).......))))... ( -50.04, z-score = -3.10, R) >droGri2.scaffold_14624 3177350 120 + 4233967 UCUCAAUGAACGUCACUAUGGCGGUUUGACGGGGCUGAAUAAGGCGGAGACCGCCGCCAAGUAUGGUGAAGCCCAGGUGCAGAUCUGGAGACGUAGCUUCGAUACACCACCGCCACCAAU ..................(((((((......(((((......(((((...)))))((((....))))..))))).((((...(((.((((......))))))).)))))))))))..... ( -43.80, z-score = -1.94, R) >anoGam1.chr2R 57986011 120 - 62725911 CCUGAACGAGCGCCACUACGGUGGCCUGACCGGGCUGAACAAGUCCGAAACGGCGGCCAAGUACGGCGAGGAGCAGGUGCUGAUCUGGCGCCGCAGCUUCGAUGUCCCCCCGCCCAACAU ........((.(((((....)))))))...((((((.....))))))....(((((((......)))..(((((.((((((.....))))))...)))))..........))))...... ( -49.00, z-score = -0.99, R) >apiMel3.GroupUn 379579214 120 - 399230636 AUUAAAUGAACGUCAUUAUGGAGGUUUGACUGGUUUAAAUAAAGCUGAAACUGCUGCUAAAUAUGGUGAAGAACAAGUUCAAAUUUGGAGAAGAUCUUUUGAUACACCUCCUCCACCUAU ....((((.....)))).((((((........(((((.....(((.......)))..)))))..((((..(((....)))..(((..(((.....)))..))).)))).))))))..... ( -22.20, z-score = -0.11, R) >triCas2.chrUn_11 411901 120 + 1365317 GCUGAACGAGCGCCAUUAUGGGGGCCUGACCGGGCUAAACAAGGCCGAAACGGCCGCCAAAUACGGCGAUGAGCAGGUGGCUAUUUGGCGGCGCAGCUUCGACGUUCCGCCCCCAGCCAU (((.....))).......(((((((..((.((((((......))))(((.(.((((((((((..(((.((......)).)))))))))))))...).)))..)).)).)))))))..... ( -48.30, z-score = -0.32, R) >consensus CCUGAACGAGCGUCACUACGGUGGACUCACUGGCCUGAACAAGGCCGAGACCGCCGCCAAGUACGGCGAGGCCCAGGUGCAGAUCUGGCGUCGCAGCUUCGACACCCCGCCACCACCGAU ...................(((((.......(((((.....)))))(((.(.((((((......))))....((((((....))))))....)).).))).........)))))...... (-29.28 = -28.15 + -1.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:33 2011