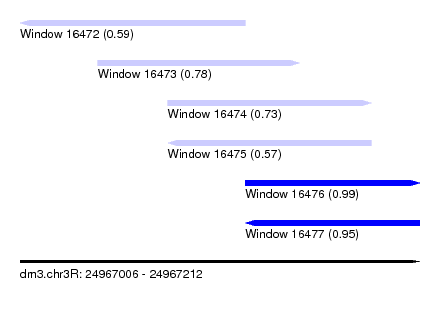
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,967,006 – 24,967,212 |
| Length | 206 |
| Max. P | 0.990909 |

| Location | 24,967,006 – 24,967,122 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.37 |
| Shannon entropy | 0.29925 |
| G+C content | 0.43734 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -23.82 |
| Energy contribution | -24.50 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.591870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24967006 116 - 27905053 CUCCGUUGGGUAGCCACUUGCAACUGGCUGGAUGGGAAGGGUGUUUCCCACUGCCAGUGUCAUCGACUGAAAUA----UAUAUUUAAAUUCGCAAUAUUUCCCAUUCAGAAGUUGUUGCU ......(((....)))...((((((..((((((((((((((.....)))..(((((((.......))))((((.----...))))......)))....))))))))))).)))))).... ( -33.60, z-score = -1.22, R) >droSim1.chr3R 24643325 116 - 27517382 CUCCGUUGGGGAGCCACUUGCAACUGUCUGGAUGGGAAGGGUGUUUCCCACUGCCAAUGUCAUCGACUGAAAUA----UAUAUUUAAAUUCGCGAUAUUUCCCAUUCAGAAGUUGUCGGU ((((.....))))..(((.((((((.(((((((((((((((.....))).......(((((..(((...((((.----...))))....))).))))))))))))))))))))))).))) ( -39.50, z-score = -2.75, R) >droSec1.super_4 3831578 116 - 6179234 CUCCGUUGGGGAGCCACUUGCAACUGGCUGGAUGGGAAGGGUGUUUCCCACUGCCAAUGUCAUCGACUGAAAUA----UAUAUUUAAAUUCGCGAUAUUUCCCAUUCAGAAGUUGUCGGU ((((.....))))..(((.((((((..((((((((((((((.....))).......(((((..(((...((((.----...))))....))).)))))))))))))))).)))))).))) ( -38.80, z-score = -2.26, R) >droEre2.scaffold_4820 7405253 99 + 10470090 -----------------CCACAUCUGGAUGGAUGGGAAGGGUGUUUCCCACAGCCAGUGUCAUCGACUGAAAUA----UAUAUUUAAAUUCGUGGUAUUUCCCAUUCUGAAGUUGUAACU -----------------..(((.((....(((((((((((.(((.....))).)).((..((.(((...((((.----...))))....)))))..)))))))))))...)).))).... ( -24.70, z-score = -0.38, R) >droYak2.chr3R 7300726 107 + 28832112 -------------CUACAUGCAACUAGCUGGAUGAAAAGGGUGUUUCCCACUGCCAGUGUCAUCGACUGAAAUAUACUUAUAUUUAAAUUCGCAAUAUUUCCCAUUCAGAAGCUGUGGCU -------------(((((.((......(((((((....(((.....)))..(((((((.......))))((((((....))))))......)))........)))))))..))))))).. ( -23.50, z-score = -0.63, R) >consensus CUCCGUUGGG_AGCCACUUGCAACUGGCUGGAUGGGAAGGGUGUUUCCCACUGCCAGUGUCAUCGACUGAAAUA____UAUAUUUAAAUUCGCGAUAUUUCCCAUUCAGAAGUUGUCGCU ...................((((((..((((((((((((((.....)))..(((.((((((...)))..(((((......)))))..))).)))....))))))))))).)))))).... (-23.82 = -24.50 + 0.68)


| Location | 24,967,046 – 24,967,150 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.04 |
| Shannon entropy | 0.25746 |
| G+C content | 0.49910 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -24.14 |
| Energy contribution | -24.70 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24967046 104 + 27905053 ----UAUAUUUCAGUCGAUGACACUGGCAGUGGGAAACACCCUUCCCAUCCAGCCAGUUGCAAGUGGCUACCCAACGGAGCUCCACUAAAAAACCCGCUAGGUGACUU------------ ----........((((..((..((((((.(((((((......)))))))...))))))..))(((((((.((....)))))..)))).....(((.....))))))).------------ ( -30.70, z-score = -1.88, R) >droSim1.chr3R 24643365 104 + 27517382 ----UAUAUUUCAGUCGAUGACAUUGGCAGUGGGAAACACCCUUCCCAUCCAGACAGUUGCAAGUGGCUCCCCAACGGAGCUCCACUAAAAAACCCGCUGGGUGACUU------------ ----.........((((((...))))))((((((((......)))))((((((...(((...(((((((((.....)))))..))))....)))...)))))).))).------------ ( -30.60, z-score = -1.26, R) >droSec1.super_4 3831618 104 + 6179234 ----UAUAUUUCAGUCGAUGACAUUGGCAGUGGGAAACACCCUUCCCAUCCAGCCAGUUGCAAGUGGCUCCCCAACGGAGCUCCACUAAAAAACCCGCUGGGUGACUU------------ ----........((((..((..((((((.(((((((......)))))))...))))))..))(((((((((.....)))))..)))).....((((...)))))))).------------ ( -34.40, z-score = -2.29, R) >droYak2.chr3R 7300766 107 - 28832112 UAAGUAUAUUUCAGUCGAUGACACUGGCAGUGGGAAACACCCUUUUCAUCCAGCUAGUUGCAUGU-------------AGAUCCACUAAAAAACCCGCUGGGUGACUUGGGUGAGCUUAG (((((.(((((((((.......))))).)))).....(((((...(((((((((..(((.....(-------------((.....)))...)))..)))))))))...))))).))))). ( -33.30, z-score = -1.94, R) >consensus ____UAUAUUUCAGUCGAUGACACUGGCAGUGGGAAACACCCUUCCCAUCCAGCCAGUUGCAAGUGGCUCCCCAACGGAGCUCCACUAAAAAACCCGCUGGGUGACUU____________ ............((((..((..((((((.(((((((......)))))))...))))))..)).(..(((((.....)))))..)........(((.....)))))))............. (-24.14 = -24.70 + 0.56)


| Location | 24,967,082 – 24,967,187 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Shannon entropy | 0.06122 |
| G+C content | 0.52381 |
| Mean single sequence MFE | -29.47 |
| Consensus MFE | -23.14 |
| Energy contribution | -24.14 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24967082 105 + 27905053 CCUUCCCAUCCAGCCAGUUGCAAGUGGCUACCCAACGGAGCUCCACUAAAAAACCCGCUAGGUGACUUAAGCUUAUAUUUAAGUCGCCGCGUAAGCCAGCCAUUA ............((.....)).(((((((.......((....))................(((((((((((......)))))))))))((....)).))))))). ( -28.50, z-score = -2.35, R) >droSim1.chr3R 24643401 105 + 27517382 CCUUCCCAUCCAGACAGUUGCAAGUGGCUCCCCAACGGAGCUCCACUAAAAAACCCGCUGGGUGACUUAAGCUUAUAGUCCAGUCGCCGCGUAAGCCAGUCAUUA ..........(((...(((...(((((((((.....)))))..))))....)))...)))(((((((...((.....))..)))))))((....))......... ( -28.70, z-score = -1.96, R) >droSec1.super_4 3831654 105 + 6179234 CCUUCCCAUCCAGCCAGUUGCAAGUGGCUCCCCAACGGAGCUCCACUAAAAAACCCGCUGGGUGACUUAAGCUUAUAUUCAAGUCGCCGCGUAAGCCAGCCAUUA ..........((((..(((...(((((((((.....)))))..))))....)))..))))((((((((............))))))))((....))......... ( -31.20, z-score = -2.81, R) >consensus CCUUCCCAUCCAGCCAGUUGCAAGUGGCUCCCCAACGGAGCUCCACUAAAAAACCCGCUGGGUGACUUAAGCUUAUAUUCAAGUCGCCGCGUAAGCCAGCCAUUA ......((((((((..(((...(((((((((.....)))))..))))....)))..))))))))......((((((..............))))))......... (-23.14 = -24.14 + 1.00)

| Location | 24,967,082 – 24,967,187 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Shannon entropy | 0.06122 |
| G+C content | 0.52381 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -34.07 |
| Energy contribution | -34.30 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.570089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24967082 105 - 27905053 UAAUGGCUGGCUUACGCGGCGACUUAAAUAUAAGCUUAAGUCACCUAGCGGGUUUUUUAGUGGAGCUCCGUUGGGUAGCCACUUGCAACUGGCUGGAUGGGAAGG ......(..(((...((((((((((((........)))))))((((((((((((((.....))))).))))))))).)))....))....)))..)......... ( -36.30, z-score = -1.47, R) >droSim1.chr3R 24643401 105 - 27517382 UAAUGACUGGCUUACGCGGCGACUGGACUAUAAGCUUAAGUCACCCAGCGGGUUUUUUAGUGGAGCUCCGUUGGGGAGCCACUUGCAACUGUCUGGAUGGGAAGG ....(((..((....(.(((((((..............)))).(((((((((((((.....))))).))))))))..))).)..))....)))............ ( -32.24, z-score = -0.08, R) >droSec1.super_4 3831654 105 - 6179234 UAAUGGCUGGCUUACGCGGCGACUUGAAUAUAAGCUUAAGUCACCCAGCGGGUUUUUUAGUGGAGCUCCGUUGGGGAGCCACUUGCAACUGGCUGGAUGGGAAGG .....((((.......))))(((((((........)))))))..(((((.((((....(((((..((((...))))..)))))...)))).)))))......... ( -38.20, z-score = -1.70, R) >consensus UAAUGGCUGGCUUACGCGGCGACUUGAAUAUAAGCUUAAGUCACCCAGCGGGUUUUUUAGUGGAGCUCCGUUGGGGAGCCACUUGCAACUGGCUGGAUGGGAAGG ......(..(((...((((((((((((........))))))).(((((((((((((.....))))).))))))))..)))....))....)))..)......... (-34.07 = -34.30 + 0.23)



| Location | 24,967,122 – 24,967,212 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 96.30 |
| Shannon entropy | 0.05102 |
| G+C content | 0.45185 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -23.12 |
| Energy contribution | -25.23 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.990909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24967122 90 + 27905053 CUCCACUAAAAAACCCGCUAGGUGACUUAAGCUUAUAUUUAAGUCGCCGCGUAAGCCAGCCAUUAAUAUUUUCCAGUCUUAGUGGGUGUG ............(((((((((((((((((((......)))))))))))((....)).......................))))))))... ( -29.80, z-score = -4.67, R) >droSim1.chr3R 24643441 90 + 27517382 CUCCACUAAAAAACCCGCUGGGUGACUUAAGCUUAUAGUCCAGUCGCCGCGUAAGCCAGUCAUUAAUAUUUUCCAGUCUUAGUGGGUGUG ............(((((((((((((((...((.....))..)))))))((....)).......................))))))))... ( -25.80, z-score = -2.88, R) >droSec1.super_4 3831694 90 + 6179234 CUCCACUAAAAAACCCGCUGGGUGACUUAAGCUUAUAUUCAAGUCGCCGCGUAAGCCAGCCAUUAAUAUUUUCCAGUCUUAGUGGGUGUG ............((((((((((((((((............))))))))((....)).......................))))))))... ( -25.40, z-score = -2.61, R) >consensus CUCCACUAAAAAACCCGCUGGGUGACUUAAGCUUAUAUUCAAGUCGCCGCGUAAGCCAGCCAUUAAUAUUUUCCAGUCUUAGUGGGUGUG ............((((((((((((((((............))))))))((....)).......................))))))))... (-23.12 = -25.23 + 2.11)


| Location | 24,967,122 – 24,967,212 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 96.30 |
| Shannon entropy | 0.05102 |
| G+C content | 0.45185 |
| Mean single sequence MFE | -24.05 |
| Consensus MFE | -24.00 |
| Energy contribution | -23.89 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.69 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24967122 90 - 27905053 CACACCCACUAAGACUGGAAAAUAUUAAUGGCUGGCUUACGCGGCGACUUAAAUAUAAGCUUAAGUCACCUAGCGGGUUUUUUAGUGGAG .....(((((((((...............((((.(((.....((.(((((((........))))))).)).))).))))))))))))).. ( -24.86, z-score = -2.10, R) >droSim1.chr3R 24643441 90 - 27517382 CACACCCACUAAGACUGGAAAAUAUUAAUGACUGGCUUACGCGGCGACUGGACUAUAAGCUUAAGUCACCCAGCGGGUUUUUUAGUGGAG .....(((((((((...............((((((((((...(((.....).)).))))))..))))((((...)))).))))))))).. ( -21.80, z-score = -0.97, R) >droSec1.super_4 3831694 90 - 6179234 CACACCCACUAAGACUGGAAAAUAUUAAUGGCUGGCUUACGCGGCGACUUGAAUAUAAGCUUAAGUCACCCAGCGGGUUUUUUAGUGGAG .....(((((((((((((............((((.......))))(((((((........)))))))..))))......))))))))).. ( -25.50, z-score = -2.01, R) >consensus CACACCCACUAAGACUGGAAAAUAUUAAUGGCUGGCUUACGCGGCGACUUGAAUAUAAGCUUAAGUCACCCAGCGGGUUUUUUAGUGGAG .....(((((((((...............((((.(((.....((.(((((((........))))))).)).))).))))))))))))).. (-24.00 = -23.89 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:31 2011