| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,026,906 – 9,027,011 |
| Length | 105 |
| Max. P | 0.594197 |

| Location | 9,026,906 – 9,027,011 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 85.55 |
| Shannon entropy | 0.30985 |
| G+C content | 0.38422 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -16.51 |
| Energy contribution | -16.35 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.594197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9026906 105 + 23011544 CCACUGUCAGCAGCACAUCA---GAUGUCU-CCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUGCCAACAGUUGUUUUGCAUCUUUGCAA ..(((((..((((....(((---(((....-..(((((((.(((((....)))))...))))))).....))))))....))))..)))))....(((((....))))) ( -27.10, z-score = -2.35, R) >droPer1.super_5 5715709 100 + 6813705 ---------CCUGGCUGUCAGCAGACAUCUUCCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUGCCAGCAGUUGUUUUGCAUCUUUGCAA ---------...((((((..((((.........(((((((.(((((....)))))...))))))).(((....)))....))))..))))))...(((((....))))) ( -24.50, z-score = -1.30, R) >dp4.chr4_group5 1308809 100 + 2436548 ---------CCUGGCUGUCAGCAGACAUCUUCCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUGCCAGCAGUUGUUUUGCAUCUUUGCAA ---------...((((((..((((.........(((((((.(((((....)))))...))))))).(((....)))....))))..))))))...(((((....))))) ( -24.50, z-score = -1.30, R) >droAna3.scaffold_12943 365544 106 + 5039921 CCGCUGUCAGCUACGCAUCCU--GACAUCU-CCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUGCCAACAGUUGUUUUGCAUCUUUGCAA ..(.((((((.........))--)))).).-..(((((((.(((((....)))))...))))))).........(((((.(((....))))))))(((((....))))) ( -21.80, z-score = -1.29, R) >droEre2.scaffold_4929 9628120 105 + 26641161 CCACUGUCAGCAGCACAUCA---GACGUCU-CCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUGCCAACAGUUGUUUUGCAUCUUUGCAA ..(((((..((((....(((---((.....-..(((((((.(((((....)))))...)))))))......)))))....))))..)))))....(((((....))))) ( -24.72, z-score = -1.77, R) >droYak2.chr2L 11686334 105 + 22324452 CCACUGUCAGCAGUACAUCA---GACGUCU-CCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUGCCAACAGUUGUUUUGCAUCUUUGCAA ..(((((..(((((...(((---((.....-..(((((((.(((((....)))))...)))))))......)))))...)))))..)))))....(((((....))))) ( -26.52, z-score = -2.62, R) >droSec1.super_3 4493397 105 + 7220098 CCACUGUCAGCAGCACAUCA---GACGUCU-CCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUGCCAACAGUUGUUUUGCAUCUUUGCAA ..(((((..((((....(((---((.....-..(((((((.(((((....)))))...)))))))......)))))....))))..)))))....(((((....))))) ( -24.72, z-score = -1.77, R) >droSim1.chr2L 8805797 105 + 22036055 CCACUGUCAGCAGCACAUCA---GACGUCU-CCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUGCCAACAGUUGUUUUGCAUCUUUGCAA ..(((((..((((....(((---((.....-..(((((((.(((((....)))))...)))))))......)))))....))))..)))))....(((((....))))) ( -24.72, z-score = -1.77, R) >droVir3.scaffold_12963 341195 107 + 20206255 CCACUGCCAUUUGGCUGGCAGCCCUCGUUU--CUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUGCCAACAGUUGUUUUGCAUCUUUGCAA ..((((((....)))((((((.........--.(((((((.(((((....)))))...))))))).(((....)))....))))))..)))....(((((....))))) ( -25.20, z-score = -1.13, R) >droMoj3.scaffold_6500 11805742 80 + 32352404 ----------------------------CU-UCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUGCCAACAGUUGUUUUGCAUCUUUGCAA ----------------------------..-..(((((((.(((((....)))))...))))))).........(((((.(((....))))))))(((((....))))) ( -16.20, z-score = -1.40, R) >droWil1.scaffold_180703 3814341 100 + 3946847 ----UGUCAGCUACAUGGC----GACAGCU-CCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUGCCAGCAGUUGUUUUGCAUCUUUGUAG ----......(((((((((----(((((..-..)))))........))))...((((((.(((...(((....)))..(((((....)))))))).))))))..))))) ( -23.30, z-score = -0.78, R) >consensus CCACUGUCAGCAGCACAUCA___GACGUCU_CCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUGCCAACAGUUGUUUUGCAUCUUUGCAA .................................(((((((.(((((....)))))...))))))).........(((((.(((....))))))))(((((....))))) (-16.51 = -16.35 + -0.16)

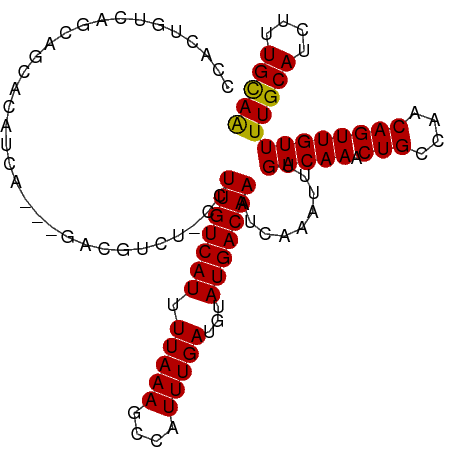
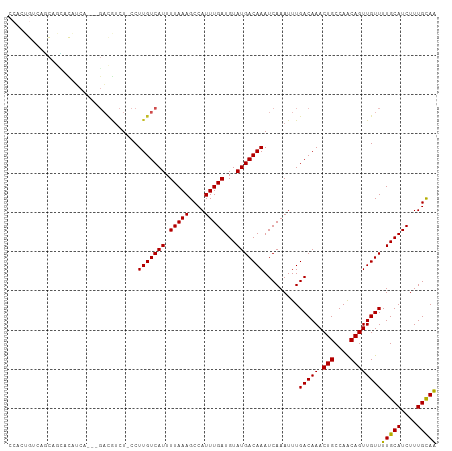
| Location | 9,026,906 – 9,027,011 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 85.55 |
| Shannon entropy | 0.30985 |
| G+C content | 0.38422 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -15.82 |
| Energy contribution | -16.00 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.517826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9026906 105 - 23011544 UUGCAAAGAUGCAAAACAACUGUUGGCAGUUUGUCAAAUUUGAUUUGUCAUACAUCAAAUGGCUUUAAAAUGACAAGG-AGACAUC---UGAUGUGCUGCUGACAGUGG (((((....)))))....(((((..(((((((((((..(((((...(((((.......))))).))))).))))))(.-...)...---......)))))..))))).. ( -33.30, z-score = -3.05, R) >droPer1.super_5 5715709 100 - 6813705 UUGCAAAGAUGCAAAACAACUGCUGGCAGUUUGUCAAAUUUGAUUUGUCAUACAUCAAAUGGCUUUAAAAUGACAAGGAAGAUGUCUGCUGACAGCCAGG--------- (((((....)))))........(((((..(((((((..(((((...(((((.......))))).))))).))))))).....((((....))))))))).--------- ( -27.70, z-score = -1.82, R) >dp4.chr4_group5 1308809 100 - 2436548 UUGCAAAGAUGCAAAACAACUGCUGGCAGUUUGUCAAAUUUGAUUUGUCAUACAUCAAAUGGCUUUAAAAUGACAAGGAAGAUGUCUGCUGACAGCCAGG--------- (((((....)))))........(((((..(((((((..(((((...(((((.......))))).))))).))))))).....((((....))))))))).--------- ( -27.70, z-score = -1.82, R) >droAna3.scaffold_12943 365544 106 - 5039921 UUGCAAAGAUGCAAAACAACUGUUGGCAGUUUGUCAAAUUUGAUUUGUCAUACAUCAAAUGGCUUUAAAAUGACAAGG-AGAUGUC--AGGAUGCGUAGCUGACAGCGG (((((....)))))..(..((((..((..(((((((..(((((...(((((.......))))).))))).))))))).-..((((.--.....)))).))..))))..) ( -26.60, z-score = -0.94, R) >droEre2.scaffold_4929 9628120 105 - 26641161 UUGCAAAGAUGCAAAACAACUGUUGGCAGUUUGUCAAAUUUGAUUUGUCAUACAUCAAAUGGCUUUAAAAUGACAAGG-AGACGUC---UGAUGUGCUGCUGACAGUGG (((((....)))))....(((((..(((((((((((..(((((...(((((.......))))).))))).))))))(.-...)...---......)))))..))))).. ( -33.30, z-score = -2.96, R) >droYak2.chr2L 11686334 105 - 22324452 UUGCAAAGAUGCAAAACAACUGUUGGCAGUUUGUCAAAUUUGAUUUGUCAUACAUCAAAUGGCUUUAAAAUGACAAGG-AGACGUC---UGAUGUACUGCUGACAGUGG (((((....)))))....(((((..(((((((((((..(((((...(((((.......))))).))))).))))))(.-...)...---......)))))..))))).. ( -33.00, z-score = -3.27, R) >droSec1.super_3 4493397 105 - 7220098 UUGCAAAGAUGCAAAACAACUGUUGGCAGUUUGUCAAAUUUGAUUUGUCAUACAUCAAAUGGCUUUAAAAUGACAAGG-AGACGUC---UGAUGUGCUGCUGACAGUGG (((((....)))))....(((((..(((((((((((..(((((...(((((.......))))).))))).))))))(.-...)...---......)))))..))))).. ( -33.30, z-score = -2.96, R) >droSim1.chr2L 8805797 105 - 22036055 UUGCAAAGAUGCAAAACAACUGUUGGCAGUUUGUCAAAUUUGAUUUGUCAUACAUCAAAUGGCUUUAAAAUGACAAGG-AGACGUC---UGAUGUGCUGCUGACAGUGG (((((....)))))....(((((..(((((((((((..(((((...(((((.......))))).))))).))))))(.-...)...---......)))))..))))).. ( -33.30, z-score = -2.96, R) >droVir3.scaffold_12963 341195 107 - 20206255 UUGCAAAGAUGCAAAACAACUGUUGGCAGUUUGUCAAAUUUGAUUUGUCAUACAUCAAAUGGCUUUAAAAUGACAAG--AAACGAGGGCUGCCAGCCAAAUGGCAGUGG (((((....)))))....(((..(((((((((((((..(((((...(((((.......))))).))))).)))))).--........)))))))(((....)))))).. ( -28.90, z-score = -1.30, R) >droMoj3.scaffold_6500 11805742 80 - 32352404 UUGCAAAGAUGCAAAACAACUGUUGGCAGUUUGUCAAAUUUGAUUUGUCAUACAUCAAAUGGCUUUAAAAUGACAAGA-AG---------------------------- (((((....)))))...(((((....)))))(((((..(((((...(((((.......))))).))))).)))))...-..---------------------------- ( -16.60, z-score = -0.92, R) >droWil1.scaffold_180703 3814341 100 - 3946847 CUACAAAGAUGCAAAACAACUGCUGGCAGUUUGUCAAAUUUGAUUUGUCAUACAUCAAAUGGCUUUAAAAUGACAAGG-AGCUGUC----GCCAUGUAGCUGACA---- (((((.....(((.......))).((((((((((((..(((((...(((((.......))))).))))).))))))..-.))))))----....)))))......---- ( -23.30, z-score = -0.47, R) >consensus UUGCAAAGAUGCAAAACAACUGUUGGCAGUUUGUCAAAUUUGAUUUGUCAUACAUCAAAUGGCUUUAAAAUGACAAGG_AGACGUC___UGAUGUGCAGCUGACAGUGG (((((....)))))...(((((....)))))(((((..(((((...(((((.......))))).))))).))))).................................. (-15.82 = -16.00 + 0.18)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:27:27 2011