
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,944,521 – 24,944,618 |
| Length | 97 |
| Max. P | 0.636872 |

| Location | 24,944,521 – 24,944,618 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 67.84 |
| Shannon entropy | 0.65842 |
| G+C content | 0.61581 |
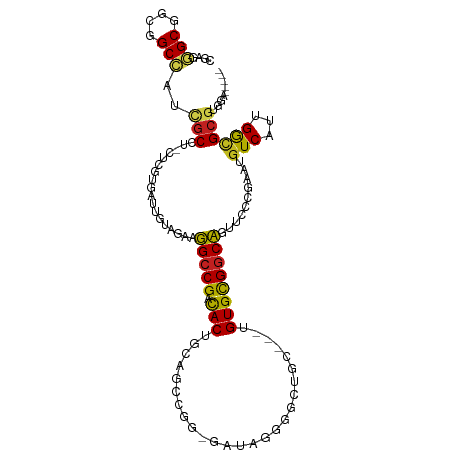
| Mean single sequence MFE | -42.80 |
| Consensus MFE | -14.43 |
| Energy contribution | -13.51 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.636872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
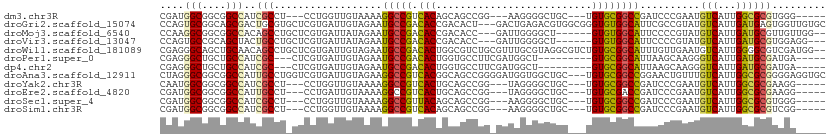
>dm3.chr3R 24944521 97 + 27905053 CGAUGGCGGCGGCCAUCGCCU---CCUGGUUGUAAAAGGCCGUCACAGCAGCCGG---AAGGGGCUGC---UGUGCGGCCGAUCCCGAAUGUCAUUGGCGCGUGGG----- .((((((....))))))(((.---...))).......(((((.((((((((((..---....))))))---)))))))))...((((..(((.....)))..))))----- ( -52.10, z-score = -2.97, R) >droGri2.scaffold_15074 6339339 108 - 7742996 CCAGUGCGGCAGCGACUGCGUGCUCGUGAUUGUAGAAUGCCGACACCGACACU---GACUGAGACGUGGCGGGUGUGGCAUUCGCCGUAUGUCAUUGAUGAGUGGUUGUGC ((((((((((.((((..(((....)))..)))).(((((((.(((((..(((.---.........)))...))))))))))))))))))).(((....))).)))...... ( -40.20, z-score = -0.52, R) >droMoj3.scaffold_6540 6165044 100 - 34148556 CCAAGGCGGCGGCCACAGCCUGCUCGUGAUUAUAGAAUGCCGACACCGACACC---GAUUGGGGCU------GUGUGGCAUUCCCCGUAUGUCAUUGAUGCGUUGUUGG-- (((..((((((((....))).((((((((((((.(((((((.((((.(...((---.....)).).------)))))))))))...))).)))).))).))))))))))-- ( -32.70, z-score = 0.34, R) >droVir3.scaffold_13047 8207742 99 + 19223366 CCAGUGCCGCAGCUACUGCCUGCUCGUGAUUAUAGAAUGCCGACACCGACACC---GAUUGGGGCU------GUGUGGCAUUCCCCGUAUGUCAUUGAUGCGUGGAGG--- (((.(((.(((((....).))))((((((((((.(((((((.((((.(...((---.....)).).------)))))))))))...))).)))).))).))))))...--- ( -30.20, z-score = 0.37, R) >droWil1.scaffold_181089 5466774 109 + 12369635 CGAGGGCAGCUGCAACAGCCUGCUCGUGAUUGUAGAAUGCCGACACUGGCGUCUGCGUUUGCGUAGGCGUCUGUGCGGCAUUUGUUGAAUGUCAUUGGGGCGUCGAUGG-- ...(((((((((...))).)))))).........((((((((.(((.((((((((((....)))))))))).)))))))))))(((((.((((.....)))))))))..-- ( -50.40, z-score = -3.28, R) >droPer1.super_0 2501121 94 + 11822988 CGAGGGCUGCUGCCAUCGC---CUCGUGAUUGUAGAAUGCCGACACUGGUGCCUUCGAUGGCU---------GUGCGGCAUUAAGCAAGGGUCAUUGAUGCGAUGA----- ....(((....)))(((((---.((((((((((..(((((((.(((....(((......))).---------))))))))))..)))...)))).))).)))))..----- ( -38.00, z-score = -1.98, R) >dp4.chr2 8687669 94 + 30794189 CGAGGGCUGCUGCCAUCGC---CUCGUGAUUGUAGAAUGCCGACACUGGUGCCUUCGAUGGCU---------GUGCGGCAUUAAGCAAGGGUCAUUGAUGCGAUGA----- ....(((....)))(((((---.((((((((((..(((((((.(((....(((......))).---------))))))))))..)))...)))).))).)))))..----- ( -38.00, z-score = -1.98, R) >droAna3.scaffold_12911 4001695 108 + 5364042 CUAGGGCGGCGGCCAUUGCCUGGUCGUGAUUGUAGAAGGCCGUCACGGCAGCCGGGGAUGGUGGCUGC---UGUGCGGCCGGAACUGUUUGUCAUUGGCGCGGGGAGGUGC ....(((....))).((.((((((((((((..(((..(((((.(((((((((((.......)))))))---)))))))))....)))...)))).)))).)))).)).... ( -49.90, z-score = -1.57, R) >droYak2.chr3R 7278231 97 - 28832112 CAAUGGCGGCGGCCAUCGCCU---CCUGGUUGUAAAAGGCCGUCACUGCAGCCGG---UAGGGGCUGC---UGUGCGGCCGAUCCCGAAUGUCAUUGGCGCGAAGG----- (((((((((((((((......---..)))))))....(((((.(((.((((((..---....))))))---.)))))))).........)))))))).........----- ( -44.80, z-score = -1.47, R) >droEre2.scaffold_4820 7382855 97 - 10470090 CGAUGGCGGCGGCCAUUGCCU---CCUGAUUGUAAAAGGCCGUCACUGCAGCCGG---UAGGGGCUGC---UGUGCGACCGAUCCCGAAUGUCAUUGGCGCGAAGG----- .((((((((.(((....))).---))...((....)).))))))((.((((((..---....))))))---.))(((.(((((..(....)..)))))))).....----- ( -36.60, z-score = 0.07, R) >droSec1.super_4 3809275 97 + 6179234 CGAUGGCGGCGGCCAUCGCCU---CCUGGUUGUAAAAGGCCGUUACAGCAGCCGG---AAGGGGCUGC---UGUGCGGCCGAUCCCGAAUGUCAUUGGCGCGUGGG----- .((((((....))))))(((.---...))).......((((((.(((((((((..---....))))))---)))))))))...((((..(((.....)))..))))----- ( -50.10, z-score = -2.62, R) >droSim1.chr3R 24620607 97 + 27517382 CGAUGGCGGCGGCCAUCGCCU---CCUGGUUGUAAAAGGCCGUCACAGCAGCCGG---AAGGGGCUGC---UGUGCGGCCGAUCCCGAAUGUCAUUGGCGCGUCGG----- .((((((....))))))(((.---...))).......(((((.((((((((((..---....))))))---)))))))))....((((.(((.....)))..))))----- ( -50.60, z-score = -2.66, R) >consensus CGAGGGCGGCGGCCAUCGCCU_CUCGUGAUUGUAGAAGGCCGACACUGCAGCCGG_GAUAGGGGCUGC___UGUGCGGCAGUUCCCGAAUGUCAUUGGCGCGUGGA_____ ....(((....)))..(((..................(((((.(((..........................))))))))..........(((...))))))......... (-14.43 = -13.51 + -0.92)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:27 2011