| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,942,420 – 24,942,531 |
| Length | 111 |
| Max. P | 0.603039 |

| Location | 24,942,420 – 24,942,531 |
|---|---|
| Length | 111 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 66.90 |
| Shannon entropy | 0.72739 |
| G+C content | 0.47934 |
| Mean single sequence MFE | -26.61 |
| Consensus MFE | -9.28 |
| Energy contribution | -8.95 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.603039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24942420 111 + 27905053 GCCAGCCGAUUCCACUGUCGCAAUUGCGUCAGAAACGGAAACAUUACUCACAGCCAGGCGGAAAGGCCCGAAAGGUU--UGCCCACCUUACACUACUCACCUUAUCUGAACCA--- (((.((.(......(((.(((....))).)))....(....)........).))..)))((...(((((....))..--.)))..))..........................--- ( -25.50, z-score = -0.71, R) >droGri2.scaffold_15074 6336990 95 - 7742996 ----GGCGACUGCAUUGUCGCACUUGUGUUAAAAAUGGCAACAUUACUCACAGUGCGACGGAAACAAGCGAAAGGUU---ACUCAUCCAACAACAGCAACAA-------------- ----.((...(((...((((((((.(((.....((((....))))...)))))))))))(....)..)))...((..---......)).......)).....-------------- ( -29.00, z-score = -3.17, R) >droMoj3.scaffold_6540 6162583 112 - 34148556 ----GACGACUGCAUUGCAGAACUUGUGUUAAAAAUGGAAACAUUACUCACAGCUCUGCCUUGGAAUCCGAAAGGUUGUUACUCAUUUGCACAAAAAAACACAUUGCACACACACA ----((((((..((..(((((.((.(((.....((((....))))...))))).)))))..)).....(....))))))).......((((.............))))........ ( -24.42, z-score = -1.30, R) >droWil1.scaffold_181089 5464150 107 + 12369635 ----GUCGCUAUCAUUGUAGAAAUUGUGUUAAAAAUGGUAACAUUACACAUAGUACUUGCGAUAAAGUCGAAAGGUAA--AUCCAUAUGAUGAUGAUGAUGAUGCAUUAGUCA--- ----((((.((((((..((.....(((((....((((....)))))))))...(((((.((((...))))..))))).--.....))..)))))).))))(((......))).--- ( -21.50, z-score = -0.41, R) >droPer1.super_0 2498838 108 + 11822988 GCGACACGCUUCCACUGUCGCAAUUGCGUCAGAAAUGGAAACAUUACUCAUAGCGCCACCGAAAGAGCCGAGAGGUC--UGCU-ACAACACUGUG--CACAGUCAUUCAAUCA--- ((((((.(......)))))))...((((.(((.((((....)))).....((((.....(....).(((....))).--.)))-).....)))))--))..............--- ( -29.10, z-score = -1.26, R) >dp4.chr2 8685385 108 + 30794189 GCAACACGCUUCCACUGUCGCAAUUGCGUCAGAAAUGGAAACAUUACUCAUAGCGCCACCGAAAGAGCCGAGAGGUC--UGCU-ACAACACUGUG--CACAGUCAUUGAAUCA--- (((...((((....(((.(((....))).))).((((....))))......))))....(....).(((....))).--))).-.(((.((((..--..))))..))).....--- ( -27.90, z-score = -0.85, R) >droAna3.scaffold_12911 3999613 108 + 5364042 GCCAGUCGAUUCCACUGUCGCAAUUGCGUCAGAAACGGAAACAUUACUCAUAGCCAGGCGGAGAGGCCAGAAAGGUU--UGCUUGCAGCUCGGG---UGCCUUAUCAAGCUUA--- (((...........(((.(((....))).)))....(....)..............)))....((((..(((((((.--.(((((.....))))---)))))).))..)))).--- ( -27.40, z-score = 1.12, R) >droYak2.chr3R 7276145 101 - 28832112 GCCAGCCGAUUCCACUGUCGCAAUUGCGUCAGAAACGGAAACAUUACUCACAGCCAGGCGGAAAGGCCCGAAAGGUU--UGCCCACC----------CACCUUAUCUAAACAA--- (((.((.(......(((.(((....))).)))....(....)........).))..)))((...(((((....))..--.)))...)----------)...............--- ( -26.30, z-score = -1.51, R) >droEre2.scaffold_4820 7380770 101 - 10470090 GCCAGCCGAUUCCACUGUCGCAAUUGCGUCAGAAACGGAAACAUUACUCACAGCCAGGCGGAAAGGCCCGAAAGGUU--UGCCCACC----------CACCUUAUCUAAGCAA--- ((..(((.......(((.(((....))).)))....(....)..............)))((...(((((....))..--.)))...)----------)...........))..--- ( -27.40, z-score = -1.43, R) >droSec1.super_4 3807190 108 + 6179234 GCAAGCCGAUUCCACUGUCGCAAUUGCGUCAGAAACGGAAACAUUACUCACAGCCAGGCGGAAAGGCCCGAAAGGUU--UGCCCAACUUACACU---CACCUUAUCUAAACCA--- (((((((..((((.(((.(((....))).)))....(....).................)))).)))((....)).)--)))............---................--- ( -27.10, z-score = -1.68, R) >droSim1.chr3R 24618509 108 + 27517382 GCAAGCCGAUUCCACUGUCGCAAUUGCGUCAGAAACGGAAACAUUACUCACAGCCAGGCGGAAAGGCCCGAAAGGUU--UGCCCAACUUACACU---CACCUUAUCUAAACCA--- (((((((..((((.(((.(((....))).)))....(....).................)))).)))((....)).)--)))............---................--- ( -27.10, z-score = -1.68, R) >consensus GCCAGCCGAUUCCACUGUCGCAAUUGCGUCAGAAACGGAAACAUUACUCACAGCCAGGCGGAAAGGCCCGAAAGGUU__UGCCCACCUCACAAU___CACCUUAUCUAAACCA___ ..............(((.(((....))).)))....(....).........................((....))......................................... ( -9.28 = -8.95 + -0.32)

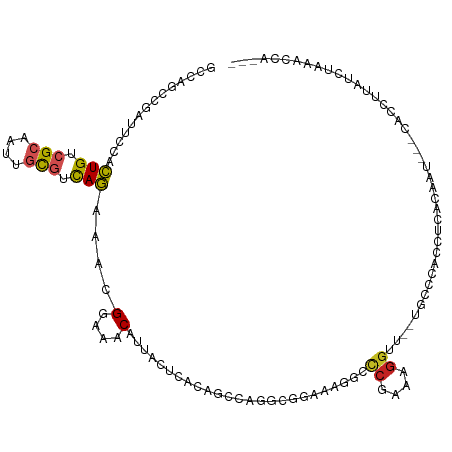

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:26 2011