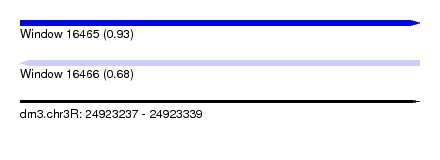
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,923,237 – 24,923,339 |
| Length | 102 |
| Max. P | 0.933313 |

| Location | 24,923,237 – 24,923,339 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 72.96 |
| Shannon entropy | 0.53806 |
| G+C content | 0.51143 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -19.82 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
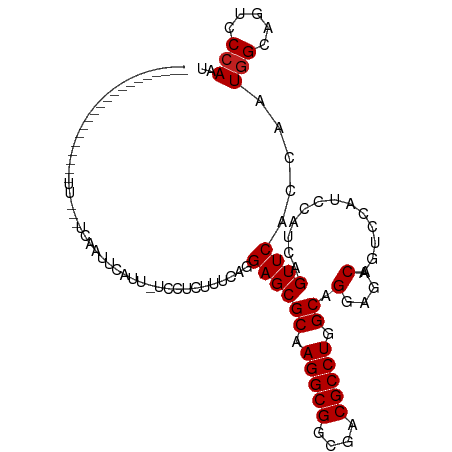
>dm3.chr3R 24923237 102 + 27905053 -----------AAUAUUAAAUUUGAUCAAUUAAUUCAUCACUUUUAGGAGCGCAAGGCGGCGACGCCUGGCAGGAGACAGUCCAUCCACCAGUUCACCAAUGGCAGUCCCAAU -----------....(((((..((((.((....)).))))..)))))((((((.(((((....))))).)).(((.........)))....)))).....(((.....))).. ( -24.10, z-score = -1.21, R) >droSim1.chr3R 24606574 102 + 27517382 -----------AAAAUUAAAUUUGGUCAAUUCAUUCUUCUGUUUCAGGAGCGCAAGGCGGCGACGCCUGGCAGGAGACAGUCCAUCCACCAGUUCACCAAUGGCAGUCCCAAU -----------..........(((((.((((.......(((((((....((.((.((((....))))))))..)))))))..........)))).)))))(((.....))).. ( -26.03, z-score = -0.63, R) >droSec1.super_4 3787939 102 + 6179234 -----------AAAAUUAAAUUUGGUCAAUUCUUUCUUCUGUUGUAGGAGCGCAAGGCGGCGACGCCUGGCAGGAGACAGUCCAUCCACCAGUUCACCAAUGGCAGUCCCAAU -----------..........((((.............((((..(..((((((.(((((....))))).)).(((.........)))....))))....)..))))..)))). ( -26.86, z-score = -0.47, R) >droEre2.scaffold_4820 7361693 101 - 10470090 -----------AAAAUUCAUUUUGUUCAAUUCAUU-UCCUCUUUUAGGAGCGCAAGGCGGCGACGCCUGGCAGGAGACAGUCCAUCCACCAGUUCACCAAUGGCAGUCCCAAU -----------.......................(-((((.....))))).((.(((((....))))).)).((.(((.(.((((..............))))).)))))... ( -24.44, z-score = -0.88, R) >droYak2.chr3R 7257111 100 - 28832112 -------------UAUUAAUUGUGUUCAAUUCAUUGUACUUUCUUAGGAGCGCAAGGCGGCGACGCCUGGCAGGAGACAGUCCAUCCACCAGUUCACCAAUGGCAGUCCCAAU -------------.....((((.(..(...((((((..((.....))((((((.(((((....))))).)).(((.........)))....))))..))))))..)..))))) ( -24.80, z-score = -0.39, R) >droAna3.scaffold_13340 4045074 93 - 23697760 -------------------UUUAUGCUUUUUUUUUUGCCAU-UUCAGGAGCGCAAGGCGGCGACGCCUGGCAGGAGACAGUCCAUCCAUCAGUUCACCAAUGGCAGCCCCAAC -------------------...............(((((((-(....((((((.(((((....))))).)).(((.........)))....))))...))))))))....... ( -30.30, z-score = -2.32, R) >dp4.chr2 21031814 79 - 30794189 ----------------------------------AAUAUUGAAAUAGGAGCGCAAGGCGGCGACGCCUGGCAGGCGACAGUCCAUCCAUCAGUUCGCAAAUGGCAGCCCCAAU ----------------------------------............((.((((.(((((....))))).)).(((....)))...((((..(....)..))))..)).))... ( -22.60, z-score = -0.06, R) >droPer1.super_0 10717354 79 + 11822988 ----------------------------------AAUCUUGAAAUAGGAGCGCAAGGCGGCGACGCCUGGCAGGCGACAGUCCAUCCAUCAGUUCGCAAAUGGCAGCCCCAAU ----------------------------------............((.((((.(((((....))))).)).(((....)))...((((..(....)..))))..)).))... ( -22.60, z-score = 0.14, R) >droWil1.scaffold_181108 4306573 87 - 4707319 --------------------------CAUUAUUUUUGCCACAAUCAGGAGCGCAAGGCGGCGACGCCUGGCAGGCGACAAUCUAUACAUCAGUUCACCAAUGGCAGUCCAAAU --------------------------........((((((.......((((((.(((((....))))).)).(....).............)))).....))))))....... ( -25.30, z-score = -2.04, R) >droVir3.scaffold_12855 9264399 85 - 10161210 ---------------------------AUCAUCUU-AUCCCAUACAGGAGCGCAAGGCGGCGACGCCUGGCAGGCGACAAUCCAUACAUCAGUUCACAAAUGGCAGUCCCAAU ---------------------------........-.(((......)))..((.(((((....))))).)).((.(((...((((..............))))..)))))... ( -22.84, z-score = -1.38, R) >droMoj3.scaffold_6540 7630286 85 + 34148556 -------------------------AUCAUUGAAU--CCUU-UGCAGGAGCGCAAGGCGGCGACGCCUGGCAGGCGACAAUCCAUACAUCAGUUCACAAAUGGCAGUCCCAAC -------------------------.........(--(((.-...))))..((.(((((....))))).)).((.(((...((((..............))))..)))))... ( -23.84, z-score = -0.73, R) >droGri2.scaffold_14906 12185308 112 - 14172833 AUCAUAAAAUAACGAACAUCAUCCGCUCUUUGCUUUGCUUU-UGCAGGAGCGCAAGGCGGCGACGCCUGGCAGGCGACAAUCCAUACAUCAGUUCACAAAUGGCAGUCCCAAU ........................((((((.((........-.))))))))((.(((((....))))).)).((.(((...((((..............))))..)))))... ( -30.84, z-score = -1.13, R) >consensus ____________________UU___UCAAUUCAUU_UCCUCUUUCAGGAGCGCAAGGCGGCGACGCCUGGCAGGAGACAGUCCAUCCAUCAGUUCACCAAUGGCAGUCCCAAU ...............................................((((((.(((((....))))).)).(....).............)))).....(((.....))).. (-19.82 = -19.90 + 0.08)

| Location | 24,923,237 – 24,923,339 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 72.96 |
| Shannon entropy | 0.53806 |
| G+C content | 0.51143 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -20.01 |
| Energy contribution | -19.42 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.46 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.679208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
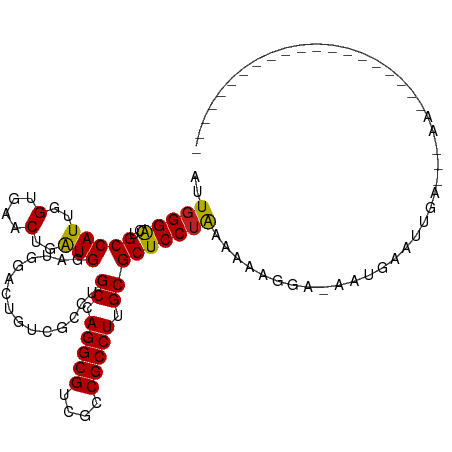
>dm3.chr3R 24923237 102 - 27905053 AUUGGGACUGCCAUUGGUGAACUGGUGGAUGGACUGUCUCCUGCCAGGCGUCGCCGCCUUGCGCUCCUAAAAGUGAUGAAUUAAUUGAUCAAAUUUAAUAUU----------- .((((((.(((...((((((.((((((((.((....))))).)))))...))))))....))).))))))...((((.((....)).))))...........----------- ( -29.70, z-score = -1.66, R) >droSim1.chr3R 24606574 102 - 27517382 AUUGGGACUGCCAUUGGUGAACUGGUGGAUGGACUGUCUCCUGCCAGGCGUCGCCGCCUUGCGCUCCUGAAACAGAAGAAUGAAUUGACCAAAUUUAAUUUU----------- .(..(((.(((...((((((.((((((((.((....))))).)))))...))))))....))).)))..).....((((.((((((.....)))))).))))----------- ( -29.10, z-score = -1.07, R) >droSec1.super_4 3787939 102 - 6179234 AUUGGGACUGCCAUUGGUGAACUGGUGGAUGGACUGUCUCCUGCCAGGCGUCGCCGCCUUGCGCUCCUACAACAGAAGAAAGAAUUGACCAAAUUUAAUUUU----------- ..(((((.(((...((((((.((((((((.((....))))).)))))...))))))....))).)))))...........((((((((......))))))))----------- ( -28.00, z-score = -0.58, R) >droEre2.scaffold_4820 7361693 101 + 10470090 AUUGGGACUGCCAUUGGUGAACUGGUGGAUGGACUGUCUCCUGCCAGGCGUCGCCGCCUUGCGCUCCUAAAAGAGGA-AAUGAAUUGAACAAAAUGAAUUUU----------- .((((((.(((...((((((.((((((((.((....))))).)))))...))))))....))).)))))).....((-(((..(((......)))..)))))----------- ( -29.00, z-score = -1.15, R) >droYak2.chr3R 7257111 100 + 28832112 AUUGGGACUGCCAUUGGUGAACUGGUGGAUGGACUGUCUCCUGCCAGGCGUCGCCGCCUUGCGCUCCUAAGAAAGUACAAUGAAUUGAACACAAUUAAUA------------- .((((((.(((...((((((.((((((((.((....))))).)))))...))))))....))).))))))............(((((....)))))....------------- ( -28.70, z-score = -0.96, R) >droAna3.scaffold_13340 4045074 93 + 23697760 GUUGGGGCUGCCAUUGGUGAACUGAUGGAUGGACUGUCUCCUGCCAGGCGUCGCCGCCUUGCGCUCCUGAA-AUGGCAAAAAAAAAAGCAUAAA------------------- ...(((((..((((..(....)..))))..(((.....))).((.(((((....))))).)))))))....-...((..........)).....------------------- ( -29.30, z-score = -0.81, R) >dp4.chr2 21031814 79 + 30794189 AUUGGGGCUGCCAUUUGCGAACUGAUGGAUGGACUGUCGCCUGCCAGGCGUCGCCGCCUUGCGCUCCUAUUUCAAUAUU---------------------------------- ...(((((..(((((........)))))..............((.(((((....))))).)))))))............---------------------------------- ( -23.30, z-score = 0.23, R) >droPer1.super_0 10717354 79 - 11822988 AUUGGGGCUGCCAUUUGCGAACUGAUGGAUGGACUGUCGCCUGCCAGGCGUCGCCGCCUUGCGCUCCUAUUUCAAGAUU---------------------------------- ...(((((..(((((........)))))..............((.(((((....))))).)))))))............---------------------------------- ( -23.30, z-score = 0.50, R) >droWil1.scaffold_181108 4306573 87 + 4707319 AUUUGGACUGCCAUUGGUGAACUGAUGUAUAGAUUGUCGCCUGCCAGGCGUCGCCGCCUUGCGCUCCUGAUUGUGGCAAAAAUAAUG-------------------------- ........((((((.(((((...(((......))).))))).((.(((((....))))).))..........)))))).........-------------------------- ( -24.00, z-score = -0.62, R) >droVir3.scaffold_12855 9264399 85 + 10161210 AUUGGGACUGCCAUUUGUGAACUGAUGUAUGGAUUGUCGCCUGCCAGGCGUCGCCGCCUUGCGCUCCUGUAUGGGAU-AAGAUGAU--------------------------- ...(((((..((((.(((......))).))))...))).)).((.(((((....))))).))..((((....)))).-........--------------------------- ( -23.00, z-score = 0.34, R) >droMoj3.scaffold_6540 7630286 85 - 34148556 GUUGGGACUGCCAUUUGUGAACUGAUGUAUGGAUUGUCGCCUGCCAGGCGUCGCCGCCUUGCGCUCCUGCA-AAGG--AUUCAAUGAU------------------------- ((((((((..((((.(((......))).))))...))).(((((.(((((....))))).))((....)).-.)))--..)))))...------------------------- ( -24.00, z-score = 0.31, R) >droGri2.scaffold_14906 12185308 112 + 14172833 AUUGGGACUGCCAUUUGUGAACUGAUGUAUGGAUUGUCGCCUGCCAGGCGUCGCCGCCUUGCGCUCCUGCA-AAAGCAAAGCAAAGAGCGGAUGAUGUUCGUUAUUUUAUGAU ...(((((..((((.(((......))).))))...))).))...(..(((((((((((((..(((..(((.-...))).))).))).)))).))))))..)............ ( -32.30, z-score = -0.04, R) >consensus AUUGGGACUGCCAUUGGUGAACUGAUGGAUGGACUGUCGCCUGCCAGGCGUCGCCGCCUUGCGCUCCUAAAAAAGGA_AAUGAAUUGA___AA____________________ ..(((((..(((((..(....)..)))...............((.(((((....))))).)))))))))............................................ (-20.01 = -19.42 + -0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:22 2011