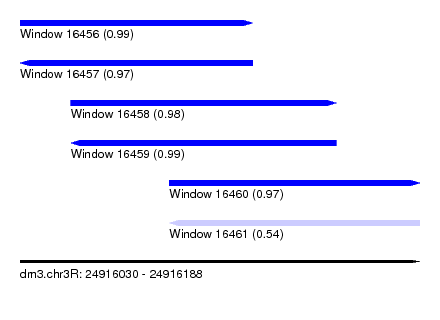
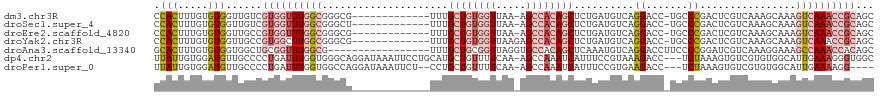
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,916,030 – 24,916,188 |
| Length | 158 |
| Max. P | 0.992191 |

| Location | 24,916,030 – 24,916,122 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 95.89 |
| Shannon entropy | 0.06182 |
| G+C content | 0.50953 |
| Mean single sequence MFE | -34.07 |
| Consensus MFE | -32.85 |
| Energy contribution | -33.35 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.990138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
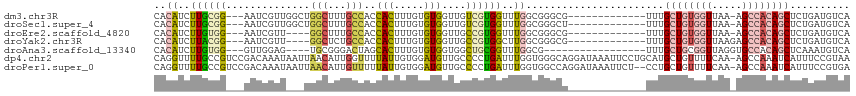
>dm3.chr3R 24916030 92 + 27905053 -AGCCGGCAAACCGCUUAUAAGCUGCGGUUUGACUUUGCUUUGACGAGUCGGGCAGGUCCUGACAUCAGAGCUGUGGCU-UUAACCACAGCAAA -.((.((((((((((.........)))))))).))..))(((((...((((((.....)))))).)))))(((((((..-....)))))))... ( -36.80, z-score = -2.90, R) >droSec1.super_4 3780837 92 + 6179234 -AGCCGGCAAACCGCUUAUAAGCUGCGGUUUGACUUUGCUUUGACGAGUCGGGCAGGUCCUGACAUCAGAGCUGUGGCU-UUAACCACAGCAAA -.((.((((((((((.........)))))))).))..))(((((...((((((.....)))))).)))))(((((((..-....)))))))... ( -36.80, z-score = -2.90, R) >droEre2.scaffold_4820 7354500 93 - 10470090 AACCCAACAUACCGCUUAUAAGCUGCGGUUUGACUUUGCUUUGACGAGUCGGGCAGGUCCUGACAUCAGAGCUGUGGCU-UUAACCACAGCAAA .......((.(((((.........))))).)).......(((((...((((((.....)))))).)))))(((((((..-....)))))))... ( -31.60, z-score = -2.56, R) >droYak2.chr3R 7249848 94 - 28832112 AACCCAACAUACCGCUUAUAAGCUGCGGUUUGACUUUGCUUUGACGAGUCGGGCAGGUCCUGACAUCAGAGCUGUGGCUCUUAACCACAGCAAA .......((.(((((.........))))).)).......(((((...((((((.....)))))).)))))(((((((.......)))))))... ( -31.10, z-score = -2.24, R) >consensus _ACCCAACAAACCGCUUAUAAGCUGCGGUUUGACUUUGCUUUGACGAGUCGGGCAGGUCCUGACAUCAGAGCUGUGGCU_UUAACCACAGCAAA .......((((((((.........)))))))).......(((((...((((((.....)))))).)))))(((((((.......)))))))... (-32.85 = -33.35 + 0.50)

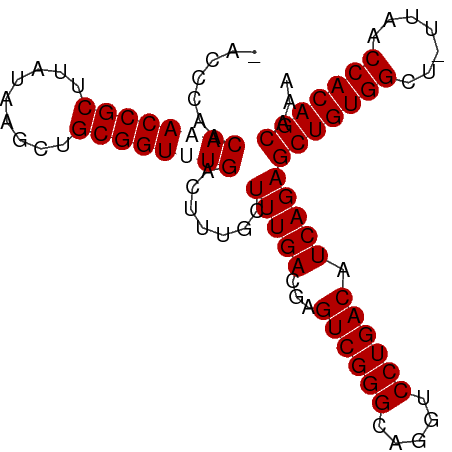
| Location | 24,916,030 – 24,916,122 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 95.89 |
| Shannon entropy | 0.06182 |
| G+C content | 0.50953 |
| Mean single sequence MFE | -33.85 |
| Consensus MFE | -29.80 |
| Energy contribution | -30.05 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24916030 92 - 27905053 UUUGCUGUGGUUAA-AGCCACAGCUCUGAUGUCAGGACCUGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGCUUAUAAGCGGUUUGCCGGCU- ...((((((((...-.))))))))..(((((...((......))....))))).(((...(.((((((((.........)))))))).).)))- ( -34.80, z-score = -2.95, R) >droSec1.super_4 3780837 92 - 6179234 UUUGCUGUGGUUAA-AGCCACAGCUCUGAUGUCAGGACCUGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGCUUAUAAGCGGUUUGCCGGCU- ...((((((((...-.))))))))..(((((...((......))....))))).(((...(.((((((((.........)))))))).).)))- ( -34.80, z-score = -2.95, R) >droEre2.scaffold_4820 7354500 93 + 10470090 UUUGCUGUGGUUAA-AGCCACAGCUCUGAUGUCAGGACCUGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGCUUAUAAGCGGUAUGUUGGGUU ...((((((((...-.))))))))((((....))))....(((((((..((....))........(((((.........)))))..))))))). ( -32.90, z-score = -2.56, R) >droYak2.chr3R 7249848 94 + 28832112 UUUGCUGUGGUUAAGAGCCACAGCUCUGAUGUCAGGACCUGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGCUUAUAAGCGGUAUGUUGGGUU ...((((((((.....))))))))((((....))))....(((((((..((....))........(((((.........)))))..))))))). ( -32.90, z-score = -2.24, R) >consensus UUUGCUGUGGUUAA_AGCCACAGCUCUGAUGUCAGGACCUGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGCUUAUAAGCGGUAUGCCGGCU_ ...((((((((.....))))))))((((....)))).((.((..((((.((....))..))))(((((((.........))))))))).))... (-29.80 = -30.05 + 0.25)

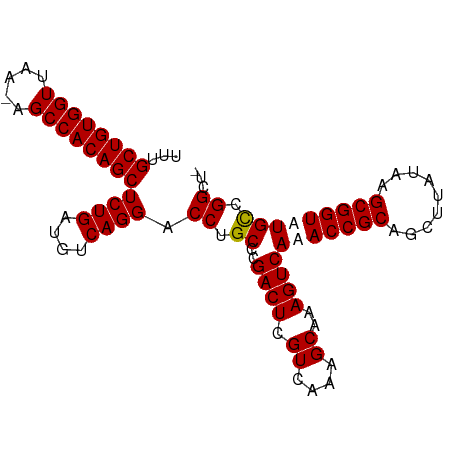
| Location | 24,916,050 – 24,916,155 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.96 |
| Shannon entropy | 0.57054 |
| G+C content | 0.52577 |
| Mean single sequence MFE | -37.91 |
| Consensus MFE | -13.27 |
| Energy contribution | -13.03 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
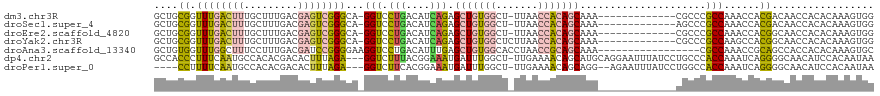
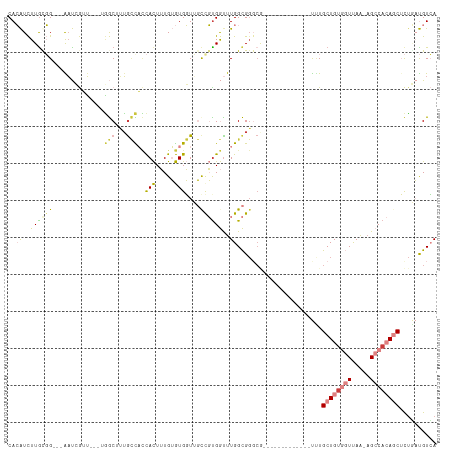
>dm3.chr3R 24916050 105 + 27905053 GCUGCGGUUUGACUUUGCUUUGACGAGUCGGGCA-GGUCCUGACAUCAGAGCUGUGGCU-UUAACCACAGCAAA-------------CGCCCGCCAAACCACGACAACCACACAAAGUGG (.((.((((((.....((((....))))(((((.-....(((....))).(((((((..-....)))))))...-------------.))))).)))))).)).)..((((.....)))) ( -38.00, z-score = -2.52, R) >droSec1.super_4 3780857 105 + 6179234 GCUGCGGUUUGACUUUGCUUUGACGAGUCGGGCA-GGUCCUGACAUCAGAGCUGUGGCU-UUAACCACAGCAAA-------------AGCCCGCCAAACCACGACAACCACACAAAGUGG (.((.((((((.(...(((((...((((((((..-...)))))).))...(((((((..-....))))))).))-------------)))..).)))))).)).)..((((.....)))) ( -38.10, z-score = -2.56, R) >droEre2.scaffold_4820 7354521 105 - 10470090 GCUGCGGUUUGACUUUGCUUUGACGAGUCGGGCA-GGUCCUGACAUCAGAGCUGUGGCU-UUAACCACAGCAAA-------------CGCCCGCCAAACCACGGCAACCACACAAAGUGG ((((.((((((.....((((....))))(((((.-....(((....))).(((((((..-....)))))))...-------------.))))).)))))).))))..((((.....)))) ( -44.60, z-score = -3.96, R) >droYak2.chr3R 7249869 106 - 28832112 GCUGCGGUUUGACUUUGCUUUGACGAGUCGGGCA-GGUCCUGACAUCAGAGCUGUGGCUCUUAACCACAGCAAA-------------CGCCCGCCAAGCCACGGCAACCACACAAAGUGG ((((.((((((.....((((....))))(((((.-....(((....))).(((((((.......)))))))...-------------.))))).)))))).))))..((((.....)))) ( -44.10, z-score = -3.18, R) >droAna3.scaffold_13340 4038571 103 - 23697760 GCUGUGGUUUGGCUUUCCUUUGACGAUCCGGGGAAGGUCCUGACAUUUGAGCUGUGGCACCUAACCGCAGCAAA-----------------CGCCAAACCGCAGCCACCACACAAAGUGC (((((((((((((((((((..(.....)..))))))..............(((((((.......)))))))...-----------------.)))))))))))))...(((.....))). ( -44.00, z-score = -3.74, R) >dp4.chr2 21025026 116 - 30794189 GCCACCCUUUCAAUGCCACACGACACUUUAGA---GGUCUUUACGGAAAUGAUUUGGCU-UUGAAAACAGCAUGCAGGAAUUUAUCCUGCCCACCAAAUCAGGGGCAACAUCCACAAUAA (((.((.........((....(((.((....)---)))).....))...((((((((((-.(....).)))..((((((.....))))))....)))))))))))).............. ( -29.10, z-score = -1.82, R) >droPer1.super_0 10710785 110 + 11822988 ----CCUUUUCAAUGCCACACGACACUUUAGA---GGUCUUCACGGAAAUGAUUUGGCU-UUGAAAACAGCAGG--AGAAUUUAUCCUGGCCACCAAAUCAGGGGCAACAUCCACAAUAA ----..(((((((.((((...(((.((....)---)))).(((......)))..)))).-)))))))...((((--(.......)))))(((.((......))))).............. ( -27.50, z-score = -1.78, R) >consensus GCUGCGGUUUGACUUUGCUUUGACGAGUCGGGCA_GGUCCUGACAUCAGAGCUGUGGCU_UUAACCACAGCAAA_____________CGCCCGCCAAACCACGGCAACCACACAAAGUGG ..........................((((((...(((.(((....))).(((((((.......))))))).....................)))...)).))))............... (-13.27 = -13.03 + -0.24)
| Location | 24,916,050 – 24,916,155 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.96 |
| Shannon entropy | 0.57054 |
| G+C content | 0.52577 |
| Mean single sequence MFE | -41.09 |
| Consensus MFE | -17.62 |
| Energy contribution | -19.49 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.992191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
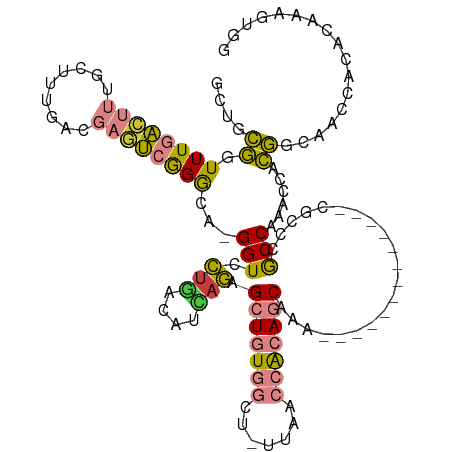
>dm3.chr3R 24916050 105 - 27905053 CCACUUUGUGUGGUUGUCGUGGUUUGGCGGGCG-------------UUUGCUGUGGUUAA-AGCCACAGCUCUGAUGUCAGGACC-UGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGC ((((.....)))).....((((((((((((((.-------------...((((((((...-.))))))))((((....))))...-.))))((....))........))))))))))... ( -47.10, z-score = -3.98, R) >droSec1.super_4 3780857 105 - 6179234 CCACUUUGUGUGGUUGUCGUGGUUUGGCGGGCU-------------UUUGCUGUGGUUAA-AGCCACAGCUCUGAUGUCAGGACC-UGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGC ((((.....)))).....((((((((((((((.-------------...((((((((...-.))))))))((((....))))...-.))))((....))........))))))))))... ( -46.60, z-score = -4.01, R) >droEre2.scaffold_4820 7354521 105 + 10470090 CCACUUUGUGUGGUUGCCGUGGUUUGGCGGGCG-------------UUUGCUGUGGUUAA-AGCCACAGCUCUGAUGUCAGGACC-UGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGC ((((.....))))..((.((((((((((((((.-------------...((((((((...-.))))))))((((....))))...-.))))((....))........)))))))))).)) ( -48.10, z-score = -4.00, R) >droYak2.chr3R 7249869 106 + 28832112 CCACUUUGUGUGGUUGCCGUGGCUUGGCGGGCG-------------UUUGCUGUGGUUAAGAGCCACAGCUCUGAUGUCAGGACC-UGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGC ((((.....))))..((.((((.(((((((((.-------------...((((((((.....))))))))((((....))))...-.))))((....))........))))).)))).)) ( -43.90, z-score = -2.20, R) >droAna3.scaffold_13340 4038571 103 + 23697760 GCACUUUGUGUGGUGGCUGCGGUUUGGCG-----------------UUUGCUGCGGUUAGGUGCCACAGCUCAAAUGUCAGGACCUUCCCCGGAUCGUCAAAGGAAAGCCAAACCACAGC .((((......))))((((.((((((((.-----------------...((((.(((.....))).)))).......((..(((..((....))..)))....))..)))))))).)))) ( -40.20, z-score = -1.64, R) >dp4.chr2 21025026 116 + 30794189 UUAUUGUGGAUGUUGCCCCUGAUUUGGUGGGCAGGAUAAAUUCCUGCAUGCUGUUUUCAA-AGCCAAAUCAUUUCCGUAAAGACC---UCUAAAGUGUCGUGUGGCAUUGAAAGGGUGGC ...........((..(((.(((((((((..((((((.....))))))....((....)).-.)))))))))((((.....((...---.))..((((((....)))))))))))))..)) ( -33.90, z-score = -1.13, R) >droPer1.super_0 10710785 110 - 11822988 UUAUUGUGGAUGUUGCCCCUGAUUUGGUGGCCAGGAUAAAUUCU--CCUGCUGUUUUCAA-AGCCAAAUCAUUUCCGUGAAGACC---UCUAAAGUGUCGUGUGGCAUUGAAAAGG---- ..............(((((......)).)))(((((.......)--))))...(((((((-.((((..((((....)))).((((---(....)).)))...)))).)))))))..---- ( -27.80, z-score = -0.96, R) >consensus CCACUUUGUGUGGUUGCCGUGGUUUGGCGGGCG_____________UUUGCUGUGGUUAA_AGCCACAGCUCUGAUGUCAGGACC_UGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGC .(((.....)))......((((((((((.....................((((((((.....))))))))..........((.......))................))))))))))... (-17.62 = -19.49 + 1.86)
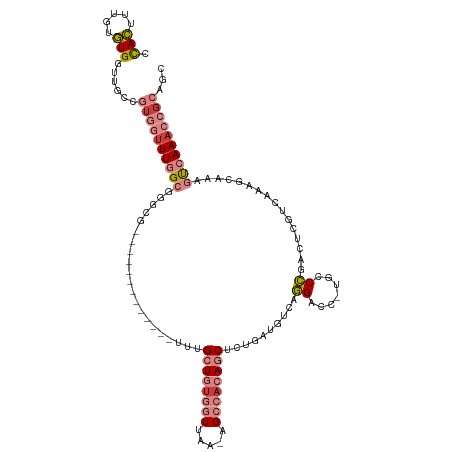
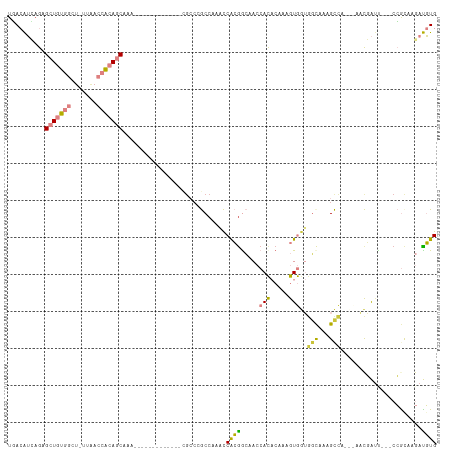
| Location | 24,916,089 – 24,916,188 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 64.48 |
| Shannon entropy | 0.62646 |
| G+C content | 0.50130 |
| Mean single sequence MFE | -29.19 |
| Consensus MFE | -9.89 |
| Energy contribution | -9.57 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.968856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24916089 99 + 27905053 UGACAUCAGAGCUGUGGCU-UUAACCACAGCAAA-------------CGCCCGCCAAACCACGACAACCACACAAAGUGGUGGCAAAGCCAGCCAACGAUU---CCGCAAGAUGUG ..(((((...(((((((..-....)))))))...-------------.....((............(((((.....)))))(((.......))).......---..))..))))). ( -30.60, z-score = -2.89, R) >droSec1.super_4 3780896 99 + 6179234 UGACAUCAGAGCUGUGGCU-UUAACCACAGCAAA-------------AGCCCGCCAAACCACGACAACCACACAAAGUGGUGGCAAAGCCAGCCAACGAUU---CCGCAAGAUGUG ..(((((...(((((((..-....)))))))...-------------.....((............(((((.....)))))(((.......))).......---..))..))))). ( -30.60, z-score = -3.07, R) >droEre2.scaffold_4820 7354560 95 - 10470090 UGACAUCAGAGCUGUGGCU-UUAACCACAGCAAA-------------CGCCCGCCAAACCACGGCAACCACACAAAGUGGUGGCAAAGCC----AACGAUU---CCACAAGAUGUG ..(((((...(((((((..-....)))))))...-------------.(((.(((.......))).(((((.....))))))))......----.......---......))))). ( -33.90, z-score = -4.03, R) >droYak2.chr3R 7249908 96 - 28832112 UGACAUCAGAGCUGUGGCUCUUAACCACAGCAAA-------------CGCCCGCCAAGCCACGGCAACCACACAAAGUGGUGGCAGAGCC----AACGAUU---CCGUAAGAUGUG ..(((((...(((((((.......)))))))...-------------.(((.(((..((....)).(((((.....)))))))).).)).----.(((...---.)))..))))). ( -34.80, z-score = -3.29, R) >droAna3.scaffold_13340 4038611 92 - 23697760 UGACAUUUGAGCUGUGGCACCUAACCGCAGCAAA-----------------CGCCAAACCGCAGCCACCACACAAAGUGCUAGUCCCGCA----CUCCAAC---CCACAAGAUGUG ..((((((..(((((((.......)))))))...-----------------.((......)).............(((((.......)))----)).....---.....)))))). ( -21.40, z-score = -1.30, R) >dp4.chr2 21025063 115 - 30794189 UUACGGAAAUGAUUUGGCU-UUGAAAACAGCAUGCAGGAAUUUAUCCUGCCCACCAAAUCAGGGGCAACAUCCACAAUAAAACCAAUGUUAAUUAUUUGUCGGACGGCAAAACCUG ....(((..((((((((((-.(....).)))..((((((.....))))))....)))))))..(....).)))......................((((((....))))))..... ( -28.30, z-score = -1.47, R) >droPer1.super_0 10710818 113 + 11822988 UCACGGAAAUGAUUUGGCU-UUGAAAACAGCAGG--AGAAUUUAUCCUGGCCACCAAAUCAGGGGCAACAUCCACAAUAAAAACAAUGUUAAUUAUUUGUCGGACGGCAAAACCUG ....((.......(((((.-(((.......((((--(.......)))))(((.((......))))).................))).)))))...((((((....)))))).)).. ( -24.70, z-score = -0.81, R) >consensus UGACAUCAGAGCUGUGGCU_UUAACCACAGCAAA_____________CGCCCGCCAAACCACGGCAACCACACAAAGUGGUGGCAAAGCCA___AACGAUU___CCGCAAGAUGUG ..........(((((((.......)))))))............................((((.....(((.....)))..(((...)))......................)))) ( -9.89 = -9.57 + -0.32)

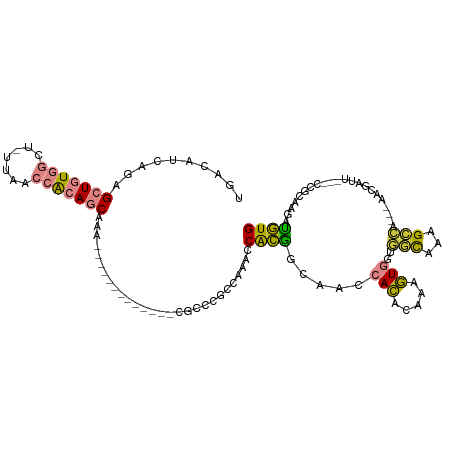
| Location | 24,916,089 – 24,916,188 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 64.48 |
| Shannon entropy | 0.62646 |
| G+C content | 0.50130 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -10.51 |
| Energy contribution | -11.50 |
| Covariance contribution | 0.99 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
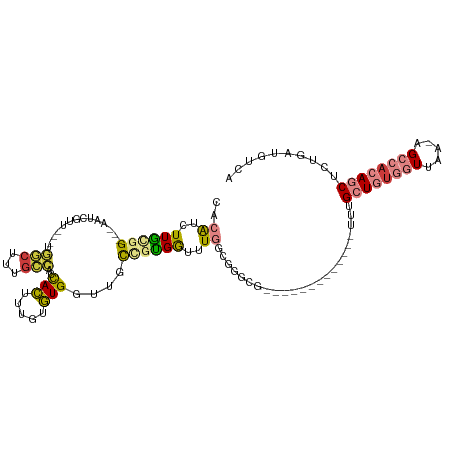
>dm3.chr3R 24916089 99 - 27905053 CACAUCUUGCGG---AAUCGUUGGCUGGCUUUGCCACCACUUUGUGUGGUUGUCGUGGUUUGGCGGGCG-------------UUUGCUGUGGUUAA-AGCCACAGCUCUGAUGUCA .......(((.(---(((((..(((.......)))(((((.....))))).....)))))).)))((((-------------((.((((((((...-.))))))))...)))))). ( -39.60, z-score = -2.51, R) >droSec1.super_4 3780896 99 - 6179234 CACAUCUUGCGG---AAUCGUUGGCUGGCUUUGCCACCACUUUGUGUGGUUGUCGUGGUUUGGCGGGCU-------------UUUGCUGUGGUUAA-AGCCACAGCUCUGAUGUCA .(((((......---.......((((.(((..((((((((.....)))))......)))..))).))))-------------...((((((((...-.))))))))...))))).. ( -38.10, z-score = -2.37, R) >droEre2.scaffold_4820 7354560 95 + 10470090 CACAUCUUGUGG---AAUCGUU----GGCUUUGCCACCACUUUGUGUGGUUGCCGUGGUUUGGCGGGCG-------------UUUGCUGUGGUUAA-AGCCACAGCUCUGAUGUCA (((((...((((---......(----(((...))))))))...)))))..(((((.....)))))((((-------------((.((((((((...-.))))))))...)))))). ( -38.90, z-score = -2.90, R) >droYak2.chr3R 7249908 96 + 28832112 CACAUCUUACGG---AAUCGUU----GGCUCUGCCACCACUUUGUGUGGUUGCCGUGGCUUGGCGGGCG-------------UUUGCUGUGGUUAAGAGCCACAGCUCUGAUGUCA .(((((.....(---((.((((----.(((..((((((((.....)))))......)))..))).))))-------------)))((((((((.....))))))))...))))).. ( -38.40, z-score = -2.10, R) >droAna3.scaffold_13340 4038611 92 + 23697760 CACAUCUUGUGG---GUUGGAG----UGCGGGACUAGCACUUUGUGUGGUGGCUGCGGUUUGGCG-----------------UUUGCUGCGGUUAGGUGCCACAGCUCAAAUGUCA .((((....(((---(((((((----(((.......)))))))..((((((((((((((......-----------------...)))))))))....))))))))))).)))).. ( -32.20, z-score = -0.48, R) >dp4.chr2 21025063 115 + 30794189 CAGGUUUUGCCGUCCGACAAAUAAUUAACAUUGGUUUUAUUGUGGAUGUUGCCCCUGAUUUGGUGGGCAGGAUAAAUUCCUGCAUGCUGUUUUCAA-AGCCAAAUCAUUUCCGUAA ..(((.....(((((.((((..(((((....)))))...)))))))))..)))..(((((((((..((((((.....))))))....((....)).-.)))))))))......... ( -30.90, z-score = -1.04, R) >droPer1.super_0 10710818 113 - 11822988 CAGGUUUUGCCGUCCGACAAAUAAUUAACAUUGUUUUUAUUGUGGAUGUUGCCCCUGAUUUGGUGGCCAGGAUAAAUUCU--CCUGCUGUUUUCAA-AGCCAAAUCAUUUCCGUGA ..(((((((.((((((...(((((..(((...))).))))).))))))..(((((......)).)))(((((.......)--)))).......)))-))))...((((....)))) ( -27.60, z-score = -1.11, R) >consensus CACAUCUUGCGG___AAUCGUU___UGGCUUUGCCACCACUUUGUGUGGUUGCCGUGGUUUGGCGGGCG_____________UUUGCUGUGGUUAA_AGCCACAGCUCUGAUGUCA ................................(((((........)))))..((((......))))...................((((((((.....)))))))).......... (-10.51 = -11.50 + 0.99)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:19 2011