| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,903,180 – 24,903,287 |
| Length | 107 |
| Max. P | 0.950514 |

| Location | 24,903,180 – 24,903,287 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 75.22 |
| Shannon entropy | 0.47267 |
| G+C content | 0.49759 |
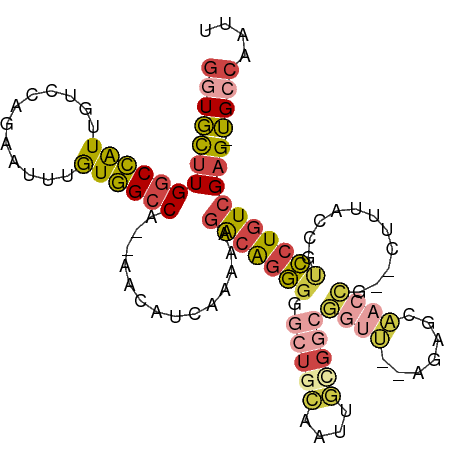
| Mean single sequence MFE | -37.77 |
| Consensus MFE | -20.08 |
| Energy contribution | -21.20 |
| Covariance contribution | 1.12 |
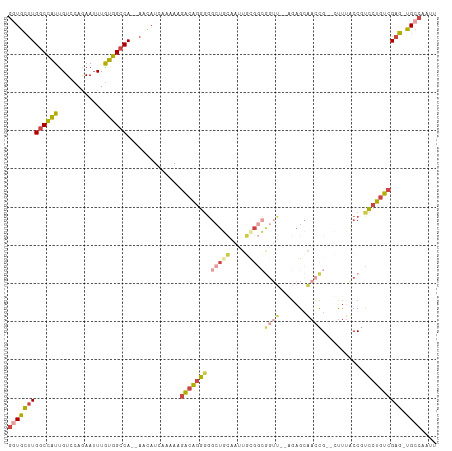
| Combinations/Pair | 1.45 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
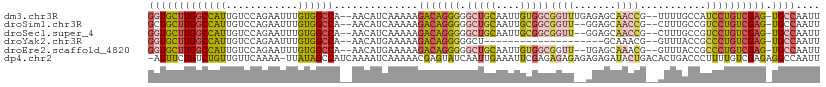
>dm3.chr3R 24903180 107 - 27905053 GGUGCUUGGCCAUUGUCCAGAAUUUGUGGCCA--AACAUCAAAAAGACAGGGGGCUGCAAUUGUGGCGGUUUGAGAGCAACCG--UUUUGCCAUCCUGUCGAG-UGCCAAUU ((..(((((((((............)))))).--...........((((((((((.((....))((((((((....).)))))--))..))).))))))))))-..)).... ( -41.50, z-score = -2.36, R) >droSim1.chr3R 24590044 105 - 27517382 GCUGCUUGGCCAUUGUCCAGAAUUUGUGGCCA--AACAUCAAAAAGACAGGGGGCUGCAAUUGCGGCGGUU--GGAGCAACCG--CUUUGCCGUCCUGUCGAG-UGCCAAUU ((.((((((((((............)))))).--...........((((((((((.((....))(((((((--(...))))))--))..))).))))))))))-)))..... ( -41.90, z-score = -1.78, R) >droSec1.super_4 3767849 105 - 6179234 GGUGCUUGGCCAUUGUCCAGAAUUUGUGGCCA--AACAUCAAAAAGACAGGGGGCUGCAAUUGCGGCGGUU--GGAGCAACCG--CUUUGCCGUCCUGUCGAG-UGCCAAUU ((..(((((((((............)))))).--...........((((((((((.((....))(((((((--(...))))))--))..))).))))))))))-..)).... ( -47.50, z-score = -3.38, R) >droYak2.chr3R 7238035 87 + 28832112 GGUGCUUGGCCAUUGUCCAGAAUUUGUGGCCA--AACAUGAAAAAGACAGGGGGCU--------------------GCAAACG--GUUUACCGCCCUGUCGAG-UGCCAAUU ((..(((((((((............)))))).--...........(((((((((..--------------------((.....--))...)).))))))))))-..)).... ( -34.60, z-score = -2.65, R) >droEre2.scaffold_4820 7342900 105 + 10470090 GGUGCUUGGCCAUUGUCCAGAAUUUGUGGCCA--AACAUGAAAAAGACAGGGGGCUGCAAUUGUGGCGGUU--UGAGCAAACG--GUUUACCGCCCUGUCGAG-UGCCAAUU ((..(((((((((............)))))).--...........(((((((.((..(....)..))(((.--.((((.....--))))))).))))))))))-..)).... ( -42.30, z-score = -2.63, R) >dp4.chr2 21012411 110 + 30794189 -AUUUCUGUCUGUUGUUCAAAA-UUAUAGCCAUCAAAAUCAAAAACGAGUAUCAAUUGAAAUUCGAGAGAGAGAGAGAUACUGACACUGACCCUUUUGUCGAGAGGCCAAUU -((((((.((((((((......-..)))))...............(((((.((....)).)))))....))).))))))..................(((....)))..... ( -18.80, z-score = 0.45, R) >consensus GGUGCUUGGCCAUUGUCCAGAAUUUGUGGCCA__AACAUCAAAAAGACAGGGGGCUGCAAUUGCGGCGGUU__AGAGCAACCG__CUUUACCGUCCUGUCGAG_UGCCAAUU (((((((((((((............))))))..............(((((((.(((((....)))))((((.......))))...........)))))))))).)))).... (-20.08 = -21.20 + 1.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:13 2011