
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,902,480 – 24,902,578 |
| Length | 98 |
| Max. P | 0.792068 |

| Location | 24,902,480 – 24,902,578 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 64.95 |
| Shannon entropy | 0.70200 |
| G+C content | 0.41372 |
| Mean single sequence MFE | -18.00 |
| Consensus MFE | -8.78 |
| Energy contribution | -9.23 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.18 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.792068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
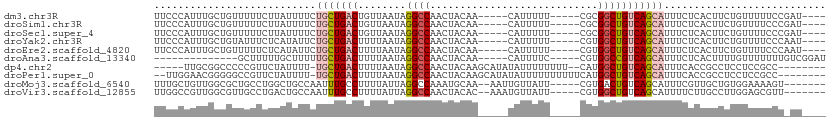
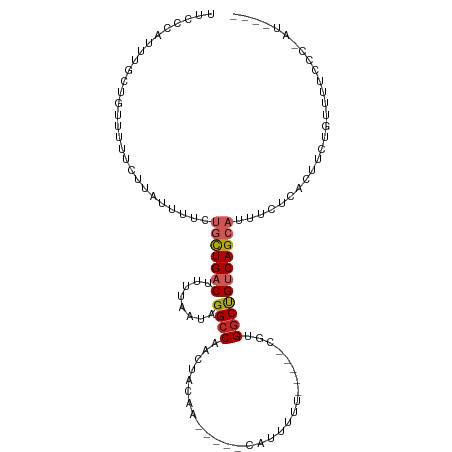
>dm3.chr3R 24902480 98 + 27905053 UUCCCAUUUGCUGUUUUUCUUAUUUUCUGCUGACUGUUAAUAGGCCAACUACAA-----CAUUUUU-----CGCGGCUGUCAGCAUUUCUCACUUCUGUUUUUCCGAU---- ...........................(((((((........((((........-----.......-----...))))))))))).......................---- ( -12.47, z-score = 0.05, R) >droSim1.chr3R 24589352 98 + 27517382 UUCCCAUUUGCUGUUUUUCUUAUUUUCUGCUGACUGUUAAUAGGCCAACUACAA-----CAUUUUU-----CGCGGCUGUCAGCAUUUCUCACUUCUGUUUUCCCGAU---- ...........................(((((((........((((........-----.......-----...))))))))))).......................---- ( -12.47, z-score = 0.06, R) >droSec1.super_4 3767145 98 + 6179234 UUCCCAUUUGCUGUUUUUCUUAUUUUCUGCUGACUGUUAAUAGGCCAACUACAA-----CAUUUUU-----CGCGGCUGUCAGCAUUUCUCACUUCUGUUUUCCCGAU---- ...........................(((((((........((((........-----.......-----...))))))))))).......................---- ( -12.47, z-score = 0.06, R) >droYak2.chr3R 7237335 98 - 28832112 UUCCCAUUUGCUGUAUUUCUCAUAUUCUGCUGACUUUUAAUAGGCCAACUACAA-----CAUUUUU-----CGUGGCUGUCAGCAUUUCUCACUUCUGUUUUCCCAAU---- ...........................(((((((........(((((.......-----.......-----..)))))))))))).......................---- ( -13.49, z-score = -1.00, R) >droEre2.scaffold_4820 7342195 98 - 10470090 UUCCCAUUUGCUGUUUUUCUCAUAUUCUGCUGACUUUUAAUAGGCCAACUACAA-----CAUUUUU-----CGUGGCUGUCAGCAUUUCUCACUUCUGUUUUCCCAAU---- ...........................(((((((........(((((.......-----.......-----..)))))))))))).......................---- ( -13.49, z-score = -1.04, R) >droAna3.scaffold_13340 4029081 88 - 23697760 --------------GCUUUUUGCUUUUUGCUGACUUUUAAUAGGCCAACUACAA-----CAUUUUC-----CGUGGCCGUCAGCAUUUCUCACUUUUGUUUUUUUGUCGGAU --------------((.....))....(((((((........(((((.......-----.......-----..))))))))))))..(((.((............)).))). ( -16.89, z-score = -1.46, R) >dp4.chr2 21011793 96 - 30794189 -----UUGCGGCCCCCGUUCUAUUUU-UGCUGACUUUUAAUAGGCCAACUACAAGCAUAUAUUUUUUUU--CAUGGCUGUCAGCAUUUCACCGCCUCCUCCGCC-------- -----..((((.....((........-(((((((........(((((......................--..)))))))))))).......)).....)))).-------- ( -18.62, z-score = -1.09, R) >droPer1.super_0 10697711 101 + 11822988 --UUGGAACGGGGGCCGUUCUAUUUU-UGCUGACUUUUAAUAGGCCAACUACAAGCAUAUAUUUUUUUUUUCAUGGCUGUCAGCAUUUCACCGCCUCCUCCGCC-------- --..(((..((((((.((........-(((((((........(((((..........................))))))))))))....)).)))))))))...-------- ( -26.87, z-score = -2.01, R) >droMoj3.scaffold_6540 7606634 98 + 34148556 UUUGCUGUUGGCGCUGCCUGGCUGCCAAUUUGCCUUUUAUUAGGCCAAAUGCAA--AAUUGUUAUU-----CGUGACUGUCAGCAUUUCGUUGCUGUGGAAAAGU------- (((((.(((((((((....))).))))))..((((......)))).....))))--).........-----.....(((.(((((......))))))))......------- ( -26.80, z-score = -0.49, R) >droVir3.scaffold_12855 9237405 98 - 10161210 UUGGCCGUUGGCGUUGCCUGACUGCCAAUUUGCCUUUUAUUAGGCCAACUACAC--AAAUGUUAUU-----CGUGGCUGUCAGCAUUUUCUUGCCUUGGAGCGUU------- ....(((..((((.(((.((((.((((....((((......)))).(((.....--....)))...-----..)))).)))))))......)))).)))......------- ( -26.40, z-score = -0.32, R) >consensus UUCCCAUUUGCUGUUUUUCUUAUUUUCUGCUGACUUUUAAUAGGCCAACUACAA_____CAUUUUU_____CGUGGCUGUCAGCAUUUCUCACUUCUGUUUUCCC_AU____ ...........................(((((((........((((............................)))))))))))........................... ( -8.78 = -9.23 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:12 2011