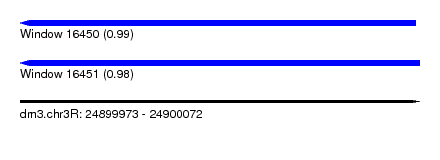
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,899,973 – 24,900,072 |
| Length | 99 |
| Max. P | 0.987396 |

| Location | 24,899,973 – 24,900,071 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 73.28 |
| Shannon entropy | 0.51728 |
| G+C content | 0.52244 |
| Mean single sequence MFE | -23.41 |
| Consensus MFE | -15.71 |
| Energy contribution | -15.71 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.987396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
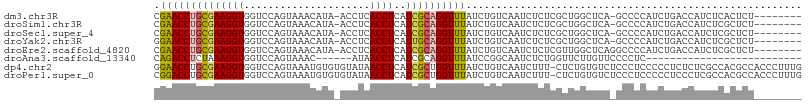
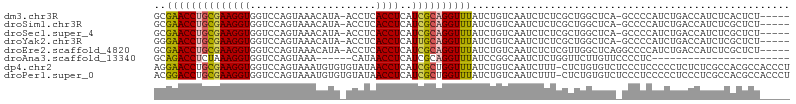
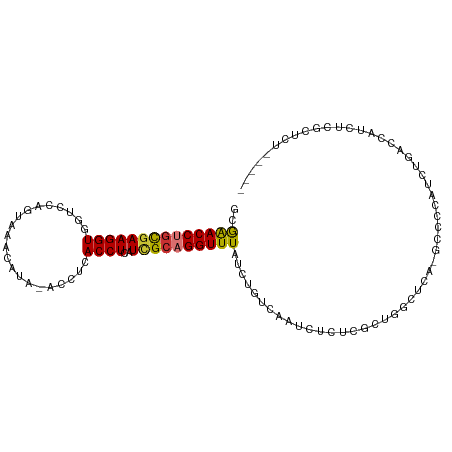
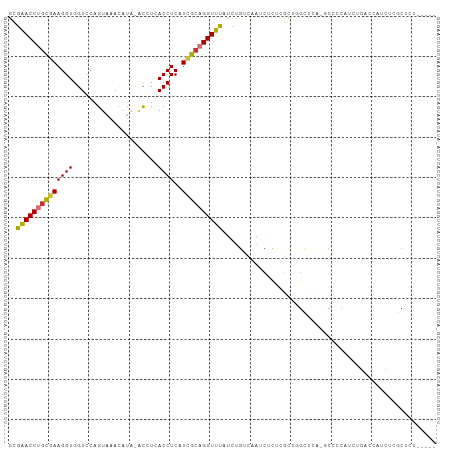
>dm3.chr3R 24899973 98 - 27905053 CGAACCUGCGAAGGUGGUCCAGUAAACAUA-ACCUCACCUCAUCGCAGGUUUAUCUGUCAAUCUCUCGCUGGCUCA-GCCCCAUCUGACCAUCUCACUCU-------- .((((((((((((((((.............-...))))))..))))))))))...............(.(((.(((-(......))))))).).......-------- ( -27.19, z-score = -3.21, R) >droSim1.chr3R 24586815 98 - 27517382 CGAACCUGCGAAGGUGGUCCAGUAAACAUA-ACCUCACCUCAUCGCAGGUUUAUCUGUCAAUCUCUCGCUGGCUCA-GCCCCAUCUGACCAUCUCGCUCU-------- .((((((((((((((((.............-...))))))..))))))))))...............(((((.(((-(......)))))))....))...-------- ( -27.59, z-score = -3.03, R) >droSec1.super_4 3764613 98 - 6179234 CGAACCUGCGAAGGUGGUCCAGUAAACAUA-ACCUCACCUCAUCGCAGGUUUAUCUGUCAAUCUCUCGCUGGCUCA-GCCCCAUCUGACCAUCUCGCUCU-------- .((((((((((((((((.............-...))))))..))))))))))...............(((((.(((-(......)))))))....))...-------- ( -27.59, z-score = -3.03, R) >droYak2.chr3R 7234766 98 + 28832112 GGAACCUGCGAAGGUGGUCCAGUAAACAUA-ACCUCACCUCAUUGCAGGUUUAUCUGUCAAUCUCUCGCUGGCUCA-GCCCCAUCUGACCAUCUCGCUCU-------- .((((((((((((((((.............-...))))))..))))))))))...............(((((.(((-(......)))))))....))...-------- ( -25.59, z-score = -1.66, R) >droEre2.scaffold_4820 7339699 99 + 10470090 CGAACCUGCGAAGGUGGUCCAGUAAACAUA-ACCUCACCUCAUCGCAGGUUUAUCUGUCAAUCUCUCGUUGGCUCAGGCCCCAUCUGACCAUCUCGCUCU-------- .((((((((((((((((.............-...))))))..))))))))))..............((.(((.(((((.....))))))))...))....-------- ( -28.59, z-score = -2.83, R) >droAna3.scaffold_13340 4027045 76 + 23697760 CAGACCUCUAAAGGUGGUCCAGUAAAC------AUAACCUCAUCGCAGGUUUAUCCGGCAAUCUCUGGUUCUUGUUCCCCUC-------------------------- ..((((.(.....).))))..(..(((------(.((((..((.((.((.....)).)).))....))))..))))..)...-------------------------- ( -14.00, z-score = -0.14, R) >dp4.chr2 21009332 107 + 30794189 GGAACCUGCGAAGGUGGUCCAGUAAAUGUGUGUAUAACCUCAUCGCUGGUUUAUCUGUCAAUCUUU-CUCUGUGUCUCCCUCCCCCUCUCUCGCCACGCCACCCUUUG ........(((((((((((((((..(((.((.....))..))).))))).................-....(((.(................).))))))).)))))) ( -17.69, z-score = -0.07, R) >droPer1.super_0 10695280 107 - 11822988 CGGACCUGCGAAGGUGGUCCAGUAAAUGUGUGUAUAACCUCAUCGCUGGUUUAUCUGUCAAUCUUU-CUCUGUGUCUCCCUCCCCCUCCCUCGCCACGCCACCCUUUG .((....((((.((.((.(((((..(((.((.....))..))).))))).......(.((......-.....)).)........)).)).))))....))........ ( -19.00, z-score = -0.24, R) >consensus CGAACCUGCGAAGGUGGUCCAGUAAACAUA_ACCUCACCUCAUCGCAGGUUUAUCUGUCAAUCUCUCGCUGGCUCA_GCCCCAUCUGACCAUCUCGCUCU________ .((((((((((((((.....................))))..))))))))))........................................................ (-15.71 = -15.71 + 0.00)
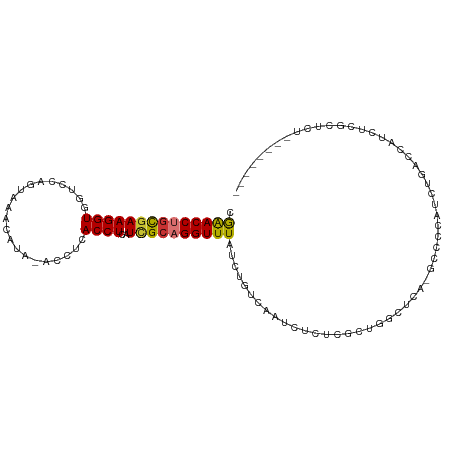
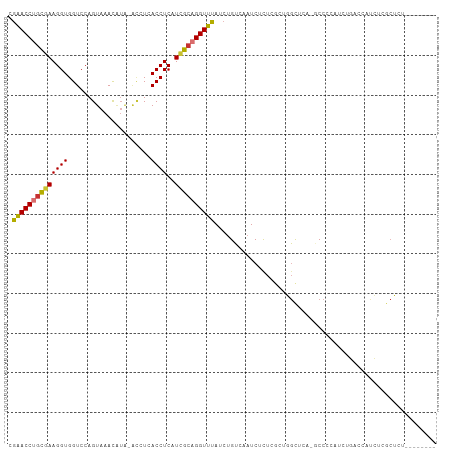
| Location | 24,899,973 – 24,900,072 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 74.02 |
| Shannon entropy | 0.51174 |
| G+C content | 0.52642 |
| Mean single sequence MFE | -24.14 |
| Consensus MFE | -15.71 |
| Energy contribution | -15.71 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.982152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24899973 99 - 27905053 GCGAACCUGCGAAGGUGGUCCAGUAAACAUA-ACCUCACCUCAUCGCAGGUUUAUCUGUCAAUCUCUCGCUGGCUCA-GCCCCAUCUGACCAUCUCACUCU----- ((((((((((((((((((.............-...))))))..))))))))...............))))(((.(((-(......))))))).........----- ( -27.35, z-score = -3.01, R) >droSim1.chr3R 24586815 99 - 27517382 GCGAACCUGCGAAGGUGGUCCAGUAAACAUA-ACCUCACCUCAUCGCAGGUUUAUCUGUCAAUCUCUCGCUGGCUCA-GCCCCAUCUGACCAUCUCGCUCU----- ((((((((((((((((((.............-...))))))..)))))))).................(.(((.(((-(......))))))).)))))...----- ( -29.59, z-score = -3.43, R) >droSec1.super_4 3764613 99 - 6179234 GCGAACCUGCGAAGGUGGUCCAGUAAACAUA-ACCUCACCUCAUCGCAGGUUUAUCUGUCAAUCUCUCGCUGGCUCA-GCCCCAUCUGACCAUCUCGCUCU----- ((((((((((((((((((.............-...))))))..)))))))).................(.(((.(((-(......))))))).)))))...----- ( -29.59, z-score = -3.43, R) >droYak2.chr3R 7234766 99 + 28832112 GGGAACCUGCGAAGGUGGUCCAGUAAACAUA-ACCUCACCUCAUUGCAGGUUUAUCUGUCAAUCUCUCGCUGGCUCA-GCCCCAUCUGACCAUCUCGCUCU----- ((((((((((((((((((.............-...))))))..))))))))...................(((.(((-(......))))))))))).....----- ( -25.79, z-score = -1.18, R) >droEre2.scaffold_4820 7339699 100 + 10470090 GCGAACCUGCGAAGGUGGUCCAGUAAACAUA-ACCUCACCUCAUCGCAGGUUUAUCUGUCAAUCUCUCGUUGGCUCAGGCCCCAUCUGACCAUCUCGCUCU----- ((((((((((((((((((.............-...))))))..)))))))).................(.(((.(((((.....)))))))).)))))...----- ( -30.59, z-score = -3.25, R) >droAna3.scaffold_13340 4027045 77 + 23697760 GCAGACCUCUAAAGGUGGUCCAGUAAA------CAUAACCUCAUCGCAGGUUUAUCCGGCAAUCUCUGGUUCUUGUUCCCCUC----------------------- ...((((.(.....).))))..(..((------((.((((..((.((.((.....)).)).))....))))..))))..)...----------------------- ( -14.00, z-score = -0.05, R) >dp4.chr2 21009335 105 + 30794189 AGGAACCUGCGAAGGUGGUCCAGUAAAUGUGUGUAUAACCUCAUCGCUGGUUUAUCUGUCAAUCUUU-CUCUGUGUCUCCCUCCCCCUCUCUCGCCACGCCACCCU .((.....((((((((((.(((((..(((.((.....))..))).))))).))))))(.((......-.....)).)..............))))....))..... ( -17.20, z-score = 0.44, R) >droPer1.super_0 10695283 105 - 11822988 ACGGACCUGCGAAGGUGGUCCAGUAAAUGUGUGUAUAACCUCAUCGCUGGUUUAUCUGUCAAUCUUU-CUCUGUGUCUCCCUCCCCCUCCCUCGCCACGCCACCCU ..((....((((.((.((.(((((..(((.((.....))..))).))))).......(.((......-.....)).)........)).)).))))....))..... ( -19.00, z-score = -0.10, R) >consensus GCGAACCUGCGAAGGUGGUCCAGUAAACAUA_ACCUCACCUCAUCGCAGGUUUAUCUGUCAAUCUCUCGCUGGCUCA_GCCCCAUCUGACCAUCUCGCUCU_____ ..((((((((((((((.....................))))..))))))))))..................................................... (-15.71 = -15.71 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:11 2011