
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,896,557 – 24,896,673 |
| Length | 116 |
| Max. P | 0.969023 |

| Location | 24,896,557 – 24,896,673 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.82 |
| Shannon entropy | 0.67634 |
| G+C content | 0.36441 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -10.84 |
| Energy contribution | -11.51 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24896557 116 + 27905053 AGCUCAUGCAUUUCUCCGCAAAGAGCGACGGGGGAAAUGCGGUGUAUCUAAUUUAGUUAUUUGGUUAUUUUCGAGCUAAAACUACUCGAAUUC----UAUAUUCUAAUUUAUAGUUUGGU (((((..((((((((((((.....))....))))))))))((.(((.((((.........)))).)))..)))))))........(((((..(----((((........)))))))))). ( -27.20, z-score = -1.35, R) >droSim1.chr3R 24583502 120 + 27517382 AGCUCAUGCAUUUCUCCGCAAAGAGCGUUGGGGGAAAUGCGGUGUAUCUAAUUUAGUUAUUUGGUUAUUUUCAAGCUAAAAGUAGUCGAAUUCUACAUAUAUUUUAAUUUAUAUUUGGGU .(((((.((((((((((.(((......)))))))))))))(((((((....((((((...((((......)))))))))).((((.......)))))))))))............))))) ( -29.80, z-score = -2.63, R) >droSec1.super_4 3761160 120 + 6179234 AGCUCAUGCAUUUCUCCGCAAAGAGCAUUGGGGGAAAUGCGGUGUAUCUUAUUUAGUUAUUUGGUUAUUUUCAAGCUAAAAGUACUCGAAUUCUACAUAUAUUUUAAUUUAUAUUUGGGU .((((((((((((((((.(((......))))))))))))))(((((....(((((((..(((((((.......)))))))...))).))))..))))).................))))) ( -29.10, z-score = -2.61, R) >droYak2.chr3R 7231349 103 - 28832112 AGCUCAUGCAUUUCUCCGCAAAAAGCGGUGGGGGAAAUGCAGUGUAUCUAAUUUAGUUAUAU--------UCGAGCUAAGAGCAAACGAAUU---------UUCUAAUUUAUAUGUGGGG (((((.(((((((((((.((........)))))))))))))(((((.(((...))).)))))--------..)))))....(((.(..((((---------....))))..).))).... ( -27.30, z-score = -2.08, R) >droEre2.scaffold_4820 7336215 103 - 10470090 AGUUCAGGCAUUUCGCCGCAAAAAGCGGUAUGGGAAAUGCAUUGUAUCUAAUUUAGUUAUUU--------UCGAGCUAAGAGUACACGAAUU---------UUCAAAUUUAUAUUUAGAU (((((..((((((((((((.....)))))....)))))))..((((.((...((((((....--------...)))))).)))))).)))))---------................... ( -23.60, z-score = -2.27, R) >droVir3.scaffold_12855 9228610 105 - 10161210 AGUCCCAGUCCUGUUUCAUACUUAGC-------GAAAUUUUACGCGUC-AGCUCAUUUUUGCA--UAACUCUGAAUAGAAGACAGGCAAGUGUGCUCUGUGACCGCAAGUACAGC----- .......(.(((((((..((.(((((-------(........)))...-.((........)).--......))).))..))))))))...((((((.(((....)))))))))..----- ( -22.30, z-score = 0.39, R) >consensus AGCUCAUGCAUUUCUCCGCAAAGAGCGGUGGGGGAAAUGCGGUGUAUCUAAUUUAGUUAUUUG__UAUUUUCGAGCUAAAAGUACUCGAAUUC____UAUAUUCUAAUUUAUAUUUGGGU .((...(((((((((((((.....))....)))))))))))..))......(((((((...............)))))))........................................ (-10.84 = -11.51 + 0.67)

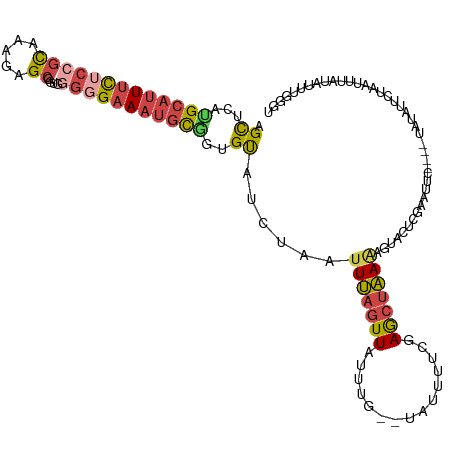
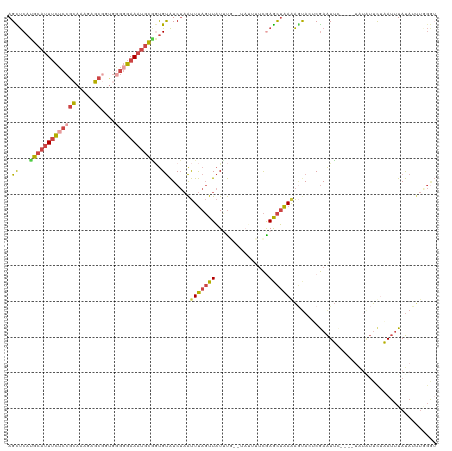
| Location | 24,896,557 – 24,896,673 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 64.82 |
| Shannon entropy | 0.67634 |
| G+C content | 0.36441 |
| Mean single sequence MFE | -20.97 |
| Consensus MFE | -7.81 |
| Energy contribution | -9.20 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.969023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24896557 116 - 27905053 ACCAAACUAUAAAUUAGAAUAUA----GAAUUCGAGUAGUUUUAGCUCGAAAAUAACCAAAUAACUAAAUUAGAUACACCGCAUUUCCCCCGUCGCUCUUUGCGGAGAAAUGCAUGAGCU ......(((((........))))----)..(((((((.......))))))).............................(((((((..((((........)))).)))))))....... ( -23.70, z-score = -2.73, R) >droSim1.chr3R 24583502 120 - 27517382 ACCCAAAUAUAAAUUAAAAUAUAUGUAGAAUUCGACUACUUUUAGCUUGAAAAUAACCAAAUAACUAAAUUAGAUACACCGCAUUUCCCCCAACGCUCUUUGCGGAGAAAUGCAUGAGCU ........................((((.......))))....(((((................(((...))).......(((((((.(((((......))).)).)))))))..))))) ( -18.30, z-score = -2.32, R) >droSec1.super_4 3761160 120 - 6179234 ACCCAAAUAUAAAUUAAAAUAUAUGUAGAAUUCGAGUACUUUUAGCUUGAAAAUAACCAAAUAACUAAAUAAGAUACACCGCAUUUCCCCCAAUGCUCUUUGCGGAGAAAUGCAUGAGCU .......................((((...(((((((.......))))))).............((.....)).))))..(((((((.(((((......))).)).)))))))....... ( -20.30, z-score = -2.49, R) >droYak2.chr3R 7231349 103 + 28832112 CCCCACAUAUAAAUUAGAA---------AAUUCGUUUGCUCUUAGCUCGA--------AUAUAACUAAAUUAGAUACACUGCAUUUCCCCCACCGCUUUUUGCGGAGAAAUGCAUGAGCU .........(((.((((..---------.(((((...((.....)).)))--------))....)))).)))....((.((((((((.....((((.....)))).)))))))))).... ( -22.40, z-score = -3.22, R) >droEre2.scaffold_4820 7336215 103 + 10470090 AUCUAAAUAUAAAUUUGAA---------AAUUCGUGUACUCUUAGCUCGA--------AAAUAACUAAAUUAGAUACAAUGCAUUUCCCAUACCGCUUUUUGCGGCGAAAUGCCUGAACU ((((((.........(...---------.)((((.((.......)).)))--------)..........)))))).....(((((((.....((((.....)))).)))))))....... ( -18.40, z-score = -2.24, R) >droVir3.scaffold_12855 9228610 105 + 10161210 -----GCUGUACUUGCGGUCACAGAGCACACUUGCCUGUCUUCUAUUCAGAGUUA--UGCAAAAAUGAGCU-GACGCGUAAAAUUUC-------GCUAAGUAUGAAACAGGACUGGGACU -----.......((.(((((((((.(((....)))))))......((((..((((--.((........)))-)))(((........)-------))......))))....))))).)).. ( -22.70, z-score = 0.89, R) >consensus ACCCAAAUAUAAAUUAGAAUAUA____GAAUUCGAGUACUUUUAGCUCGAAAAUA__CAAAUAACUAAAUUAGAUACACCGCAUUUCCCCCAACGCUCUUUGCGGAGAAAUGCAUGAGCU ...........................................(((((................((.....)).......(((((((((.((........)).)).)))))))..))))) ( -7.81 = -9.20 + 1.39)

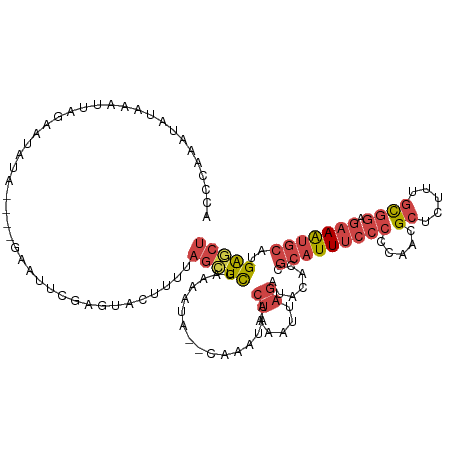
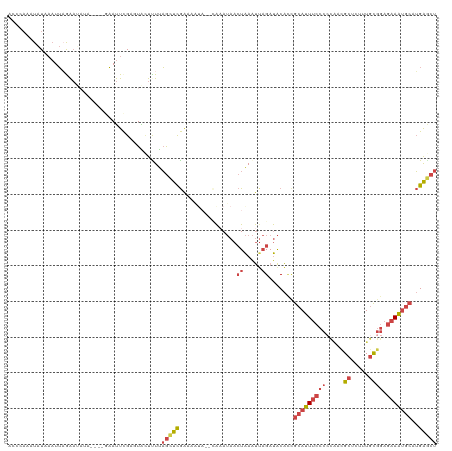
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:08 2011