
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,890,260 – 24,890,355 |
| Length | 95 |
| Max. P | 0.534047 |

| Location | 24,890,260 – 24,890,355 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.67 |
| Shannon entropy | 0.51205 |
| G+C content | 0.41596 |
| Mean single sequence MFE | -18.28 |
| Consensus MFE | -9.73 |
| Energy contribution | -9.72 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.534047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24890260 95 + 27905053 -----------AAAAAACACCACACAACAGAAGAGCAGCAC-AAAUCUUUUGCCAAUGAGUCGCAGACAGUCAUUGGAUCGACCAGGUGAGAAAAUGCCA-AUCAAAA--------- -----------......((((......(((((((.......-...)))))))((((((((((...)))..)))))))........))))...........-.......--------- ( -20.20, z-score = -1.97, R) >droGri2.scaffold_14906 12144506 83 - 14172833 -------------------AGCAUAAA-ACAAGG--CGUAU-AAAU-UUUUGCCAAUGAGUCGCAGACAGUCAUUGGAUCGACCAGGUGAGAAU-AUAUAAAAAAAAA--------- -------------------.((.....-.....)--).(((-(.((-(((..((..((.((((.(..(((...)))..)))))))))..)))))-.))))........--------- ( -15.30, z-score = -1.64, R) >droMoj3.scaffold_6540 7591631 83 + 34148556 -------------------AGCAUAAA-ACAAGG--CGCAU-AAAU-UUUUGCCAAUGAGUCGCAGACAGUCAUUGGAUCGACCAGGUGAGAAU-ACAAAAAAAAAAA--------- -------------------.((.....-.....)--)....-..((-(((..((..((.((((.(..(((...)))..)))))))))..)))))-.............--------- ( -13.30, z-score = -0.52, R) >droVir3.scaffold_12855 9219437 84 - 10161210 -------------------AGCAUUAA-ACAAGG--CUCAA-AAAU-UUUUGCCAAUGAGUCGCAGACAGUCAUUGGAUCGACCAGGUGAGCAUUACAAAAAAAAAAA--------- -------------------........-.....(--((((.-....-.....((((((((((...)))..)))))))..........)))))................--------- ( -17.45, z-score = -1.70, R) >droWil1.scaffold_181108 4281765 101 - 4707319 ----ACCCUCCCCCCAAAGAAACGCAACACAAGG--CGCAUCAAAUAUUUUGCCAAUGAGUCGCAGACAGUCAUUGGAUCGACCAGGUGAGUGA-AAAUAAGUGAAAA--------- ----.(.(((((......((..(((........)--))..))..........((((((((((...)))..)))))))........)).))).).-.............--------- ( -17.60, z-score = 0.20, R) >dp4.chr2 21001192 108 - 30794189 ACACCACCCUCUACAACAAAACAGCAGCACAGGG--AGCAU-AAAUCUUUUGCCAAUGAGUCGCAGACAGUCAUUGGAUCGACCAGGUGAGUGU-AUAGAAAAAAAAAAAAA----- ((((((((..................(((.((((--(....-...))))))))(((((((((...)))..)))))).........)))).))))-.................----- ( -20.40, z-score = -0.58, R) >droPer1.super_0 10687124 104 + 11822988 ACACCACCCUCUACAACAAAACAGCAGCACAGGG--AGCAU-AAAUCUUUUGCCAAUGAGUCGCAGACAGUCAUUGGAUCGACCAGGUGAGUGU-AUAGAAAAAAAAA--------- ((((((((..................(((.((((--(....-...))))))))(((((((((...)))..)))))).........)))).))))-.............--------- ( -20.40, z-score = -0.42, R) >droSim1.chr3R 24577344 95 + 27517382 -----------AAAACACACCACACAACAGAAGAGCAGCAC-AAAUCUUUUGCCAAUGAGUCGCAGACAGUCAUUGGAUCGACCAGGUGAGAAAAUGCCA-AUCAAAA--------- -----------......((((......(((((((.......-...)))))))((((((((((...)))..)))))))........))))...........-.......--------- ( -20.20, z-score = -1.86, R) >droSec1.super_4 3754964 95 + 6179234 -----------AAAACACACCACACAACAGAAGAGCAGCAC-AAAUCUUUUGCCAAUGAGUCGCAGACAGUCAUUGGAUCGACCAGGUGAGAAAAUGCCA-AUCAAAA--------- -----------......((((......(((((((.......-...)))))))((((((((((...)))..)))))))........))))...........-.......--------- ( -20.20, z-score = -1.86, R) >droYak2.chr3R 7224943 95 - 28832112 -----------AAAAAACACCACACAACAGAAGAGCAGCAC-AAAUCUUUUGCCAAUGAGUCGCAGACAGUCAUUGGAUCGACCAGGUGAGAAAAUGCCA-AUCACAA--------- -----------......((((......(((((((.......-...)))))))((((((((((...)))..)))))))........))))...........-.......--------- ( -20.20, z-score = -1.86, R) >droEre2.scaffold_4820 7329955 95 - 10470090 -----------AAAAAACACCACACAACAGAAGAGCAGCAC-AAAUCUUUUGCCAAUGAGUCGCAGACAGUCAUUGGAUCGACCAGGUGAGAAAAUGCCA-AUCACAC--------- -----------......((((......(((((((.......-...)))))))((((((((((...)))..)))))))........))))...........-.......--------- ( -20.20, z-score = -1.89, R) >droAna3.scaffold_13340 4019553 103 - 23697760 -----------AAACCACAACACACAGCACAAGG--AGCCC-AAAUCUUUUGCCAAUGAGUCGCAGACAGUCAUUGGAUCGACCAGGUGAGAACAAAUGACAUCAAACCGAAUAAAA -----------..(((..........(((.((((--(....-...))))))))(((((((((...)))..)))))).........)))............................. ( -13.90, z-score = 0.77, R) >consensus ___________AAAAAACAACACACAACACAAGG__AGCAC_AAAUCUUUUGCCAAUGAGUCGCAGACAGUCAUUGGAUCGACCAGGUGAGAAAAAGACAAAUCAAAA_________ ................................................((..((..((.((((.(..(((...)))..)))))))))..)).......................... ( -9.73 = -9.72 + -0.00)

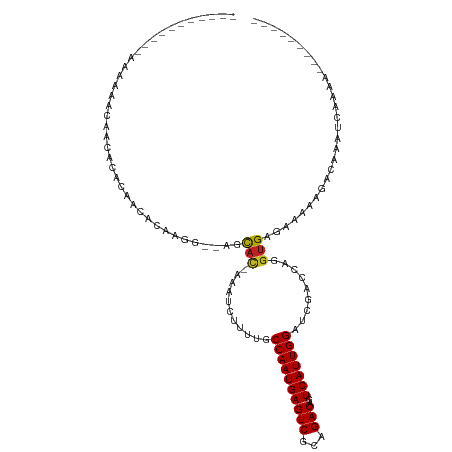
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:05 2011