| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,869,663 – 24,869,768 |
| Length | 105 |
| Max. P | 0.971789 |

| Location | 24,869,663 – 24,869,768 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 71.82 |
| Shannon entropy | 0.50953 |
| G+C content | 0.48400 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -12.78 |
| Energy contribution | -12.46 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.971789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
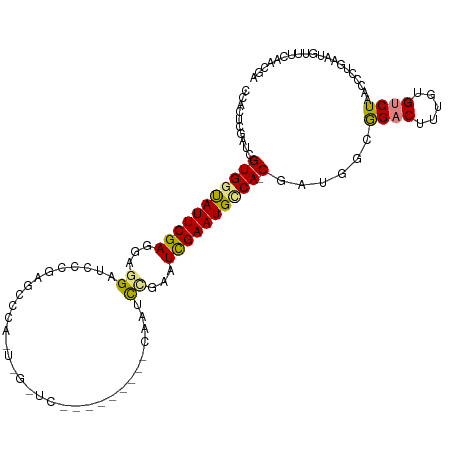
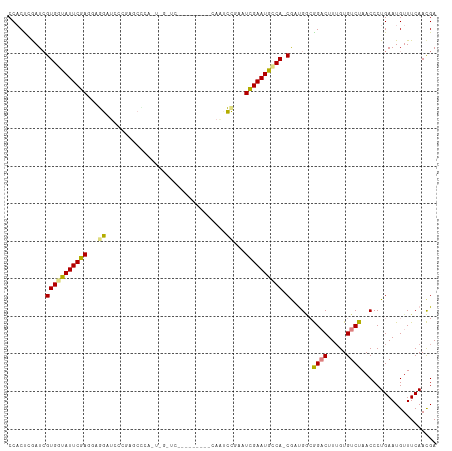
>dm3.chr3R 24869663 105 + 27905053 CCACCAGAUCGUGGCAUUCGAGGAGGAUCACGAGCCCAUUUGCUCU--------CAAUCCGAGUCGAAUGUCA-CAAUGGCGGACUUUGUGUCUAACCCUGAAUGUUUCAACGA ....(((...(((((((((((...((((...((((......)))).--------..))))...))))))))))-)...((.((((.....))))..)))))............. ( -33.90, z-score = -1.70, R) >droSim1.chr3R 24556147 105 + 27517382 CCACUAGAUCGUGGCAUUCGAGAAGGAUCCCGAGCCGAAUUGCUCC--------CAAUCCGAGUCGAAUGCCA-CGAUGGCGGACUUUGUGUCUAACCCUGAAUGUUUCAACGA ....(((((((((((((((((...((((...((((......)))).--------..))))...))))))))))-))))((.((((.....))))..)))))............. ( -39.00, z-score = -3.81, R) >droSec1.super_4 3732764 105 + 6179234 CCACUAGAUCGUGGCAUUCGAGAAGGAUCCCGAGCCCAAUUGCUCC--------CAAUCCGAGUCGAAUGCCA-CGUUGCCGGACUUUGUGUCUAACCCUGAAUGUUUCAACGA ....(((..((((((((((((...((((...((((......)))).--------..))))...))))))))))-)).....((((.....))))....)))............. ( -35.10, z-score = -3.47, R) >droYak2.chr3R 7203003 112 - 28832112 CCACCCAAUCGUGGUAUUCGAUCAGGAUCCUUGGCUCCUUGGAUCCUUGGAUACCAAUCCAAAUCGAAUGCCA-CGAUGGCGGACUU-GUGUCUAACCCUGAAUGUUUCAACGA ....(((.((((((((((((((..((((((..(....)..))))))((((((....)))))))))))))))))-)))))).((((..-..)))).................... ( -45.20, z-score = -5.25, R) >droEre2.scaffold_4820 7306026 90 - 10470090 CCACUCGAUCGUGGUAUUCGAGCAGGAUACU-----------------------CAGCCUAAAUCGAAUGCCA-CGAUGGCGGACUUUGUGUCUAACCCUGAAUGUUUCAACGA ....(((((((((((((((((..(((.....-----------------------...)))...))))))))))-))))((.((((.....))))...))............))) ( -30.90, z-score = -3.20, R) >dp4.chr2 17631310 97 - 30794189 CCUCUCAAUGGUGAUAUUCGAUGGAGAGGAGAGUCUAA-----------------AAAUCGCAUUGAAUGCCAUCAUCGAGAGCCUUUGUGUCUAACCCUGAAUGUUUCAAUGA .(((((.((((((.(((((((((((.(((....)))..-----------------...)).))))))))).)))))).)))))............................... ( -24.00, z-score = -0.81, R) >droPer1.super_0 7289293 97 + 11822988 CCUCUCAAUGGUGAUAUUCGAUGGAGAGGAGAGUGCUA-----------------AAAUCGCAUUGAAUGCCAUCAUCGAGAGCCUUUGUGUCUAACCCUGAAUGUUUCAAUGA .(((((.((((((.(((((((((((....((....)).-----------------...)).))))))))).)))))).)))))............................... ( -25.20, z-score = -1.04, R) >consensus CCACUCGAUCGUGGUAUUCGAGGAGGAUCCCGAGCCCA_U_G_UC_________CAAUCCGAAUCGAAUGCCA_CGAUGGCGGACUUUGUGUCUAACCCUGAAUGUUUCAACGA ........(((((((((((((...((................................))...)))))))))..((..(..((((.....))))..)..)).........)))) (-12.78 = -12.46 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:03 2011