| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,840,665 – 24,840,794 |
| Length | 129 |
| Max. P | 0.831461 |

| Location | 24,840,665 – 24,840,794 |
|---|---|
| Length | 129 |
| Sequences | 7 |
| Columns | 131 |
| Reading direction | reverse |
| Mean pairwise identity | 92.14 |
| Shannon entropy | 0.15734 |
| G+C content | 0.33998 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -22.12 |
| Energy contribution | -22.33 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.831461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
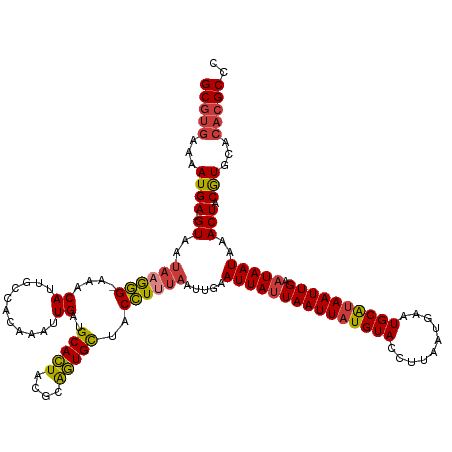
>dm3.chr3R 24840665 129 - 27905053 GCGUGAAAAUGA-GUAUUAUGGG-AAACAUUGCCACAAAUUGAUGCACUACGCAGUGCUACCUUUAAUUGAAUUAUUAAUUAUGUACCUUAAUGAAUGCAUAAUUGAAUAAUAAACUACGUGCACACGCCC (((((.......-........(.-...)..(((.((.((((((.(((((....))))).....))))))..(((((((((((((((..........)))))))))).))))).......)))))))))).. ( -27.80, z-score = -2.01, R) >droSim1.chr3R 24527259 129 - 27517382 GCGUGAAAAUGA-GUAAUAAGGG-AAACAUUGCCACAAAUUGAUGCACUACGCAGUGCUACCUUUAAUUGAAUUAUUAAUUAUGUACCUUAAUGAAUGCAUAAUUGAAUAAUAAACUACGUGCACACGCCC (((((...((((-((..((((((-...((((((..................))))))...)))))).....(((((((((((((((..........)))))))))).)))))..))).)))...))))).. ( -28.97, z-score = -2.60, R) >droSec1.super_4 3702723 129 - 6179234 GCGUGAAAAUGA-GUAAUAAGGG-AAACAUUGCCACAAAUUGAUGCACUACGCAGUGCUACCUUUAAUUGAAUUAUUAAUUAUGUACCUUAAUGAAUGCAUAAUUGAAUAAUAAACUACGUGCACACGCCC (((((...((((-((..((((((-...((((((..................))))))...)))))).....(((((((((((((((..........)))))))))).)))))..))).)))...))))).. ( -28.97, z-score = -2.60, R) >droYak2.chr3R 7173799 129 + 28832112 GCGUGAAAAUGA-GUAAUAAGGG-AAACAUUGCCACAAAUUGAUGCACUACGCAGUGCUACCUUUAAUUGAAUUAUUAAUUAUGUACCUUAAUGAAUGCGUAAUUGAAUAAUAAACUACGUGCACACGCCC (((((...((((-((..((((((-...((((((..................))))))...)))))).....(((((((((((((((..........)))))))))).)))))..))).)))...))))).. ( -28.57, z-score = -2.14, R) >droEre2.scaffold_4820 7276858 129 + 10470090 GCGUGAAAAUGA-GUAAUAAGGG-AAACAUUGCCACAAAUUGAUGCACUACGCAGUGCUACCUUUAAUUGAAUUAUUAAUUAUGUACCUUAAUGAAUGCGUAAUUGAAUAAUAAACUACGUGCACACGCCC (((((...((((-((..((((((-...((((((..................))))))...)))))).....(((((((((((((((..........)))))))))).)))))..))).)))...))))).. ( -28.57, z-score = -2.14, R) >droPer1.super_0 7260664 129 - 11822988 GCGUGAAAAUGA-GUAAUAAAGG-AAACAUUGCCACAAAUUGAUGCACUACGCGGUGCUACCUUUAAUUCAAUUAUUAAUUAUGUACCUUAAUGAAUGCAUAAUUGAAUAAUAAACUACGUGCACACGCCU (((((...((((-((..((((((-...((...........))..(((((....)))))..))))))((((((((((((.((((........)))).)).)))))))))).....))).)))...))))).. ( -29.20, z-score = -2.73, R) >droMoj3.scaffold_6540 20693883 129 + 34148556 GCAAAAAAAUGACGUAAUUUAAGUGAACAGAAAAUCAAAUUGAGGCAAUA-AUGUUGUUACUUUU-AUUGAAUUAAUAAUUCUGUACCUUAAUGAAUGCAUAAUUGAAUAAUAAACUACAUGCACACGCCC ((................(((((.(.((((((......(((((..(((((-(.((....))..))-))))..)))))..)))))).))))))....(((((..................)))))...)).. ( -18.17, z-score = -0.25, R) >consensus GCGUGAAAAUGA_GUAAUAAGGG_AAACAUUGCCACAAAUUGAUGCACUACGCAGUGCUACCUUUAAUUGAAUUAUUAAUUAUGUACCUUAAUGAAUGCAUAAUUGAAUAAUAAACUACGUGCACACGCCC (((((...(((......((((((....((((((..................))))))...)))))).....(((((((((((((((..........)))))))))).)))))......)))...))))).. (-22.12 = -22.33 + 0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:53:02 2011