
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,806,844 – 24,806,939 |
| Length | 95 |
| Max. P | 0.558954 |

| Location | 24,806,844 – 24,806,939 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 71.86 |
| Shannon entropy | 0.54543 |
| G+C content | 0.49839 |
| Mean single sequence MFE | -31.04 |
| Consensus MFE | -7.74 |
| Energy contribution | -8.23 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.558954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24806844 95 + 27905053 ------GCAGCUUAUUGGCCUUGGCUCCUUCGAUUGCGGCUAUCAAAUCAUCGGUACAACGUCAUUAACAUGCC-GCC--AAUGAAGC--AGUCAAGCCGAAAGCU-------- ------..(((((.(((((.((((((.(((((...(((((...........((......))..........)))-)).--..))))).--))))))))))))))))-------- ( -34.70, z-score = -3.75, R) >droSim1.chr3R 24490713 101 + 27517382 GCUGGUGCGGCUUAUUGGCCCUGGCUCCUUCGAUUGCGGCUAUCAAAUCAUCGGUACAACGUCAUUAACAUGCC-GCC--AAUGAAGC--AGUCAAGCCGAAAGCU-------- ((....))(((((.(((((..(((((.(((((...(((((...........((......))..........)))-)).--..))))).--))))).))))))))))-------- ( -34.50, z-score = -1.81, R) >droSec1.super_4 3669291 101 + 6179234 GCUGGUGCGGCUUAUUGGCCCUGGCUCCUUCGAUUGCGGCUAUCAAAUCAUCGGUACAACGUCAUUAACAUGCC-GCC--AAUGAAGC--AGUCAAGCCGAAAGCU-------- ((....))(((((.(((((..(((((.(((((...(((((...........((......))..........)))-)).--..))))).--))))).))))))))))-------- ( -34.50, z-score = -1.81, R) >droYak2.chr3R 7139362 98 - 28832112 ------GCGGCUUAUUGGCCCUUGCUCCUUCGAUUGCGGCUAUCAAAUCAUCGGUACAACGUCAUUAACAUGCC-GCC--AAUGAAGC--AGUCAAGCCGAAAGCUGAA----- ------.((((((.(((((..(((((.(((((...(((((...........((......))..........)))-)).--..))))).--)).))))))))))))))..----- ( -31.60, z-score = -2.29, R) >droEre2.scaffold_4820 7240176 95 - 10470090 ------CCGGCUUAUUGGCCCUGGCUCCUUCGAUUGCGGCUAUCAAAUCAUCGGUACAACGUCAUUAACAUGCC-GCC--AAUGAAGC--AGUCAAGCCGAAAGCU-------- ------..(((((.(((((..(((((.(((((...(((((...........((......))..........)))-)).--..))))).--))))).))))))))))-------- ( -33.00, z-score = -3.05, R) >droAna3.scaffold_13340 3935703 90 - 23697760 ------CCGGCUUAUUGGC------UGCUUCGAAUGCGGCUAUCAAAUCAUCGGUACAACGCCAUUAACAUGCCCGCC--AAUGAAGC--AGUCAAGCCGAAAGCU-------- ------.(((((...((((------(((((((...((((.(((........((......))........))).)))).--..))))))--))))))))))......-------- ( -31.99, z-score = -3.06, R) >dp4.chr2 17565083 94 - 30794189 ---AUUCUGGCUUAUUGGC------UUCUCCGACUGCGGCUAUCAAAUCAGCGGUACAACGUCAUUAACAUGUUGGCCC-AAUGAAGC--AGUCAAGCUGAAAGCC-------- ---.....(((((.(..((------((....((((((.(((........)))((..((((((.......))))))..))-......))--))))))))..))))))-------- ( -32.30, z-score = -3.13, R) >droPer1.super_0 7224175 94 + 11822988 ---AUUCUGGCUUAUUGGC------UUCUCCGACUGCGGCUAUCAAAUCAGCGGUACAACGUCAUUAACAUGUUGGCCC-AAUGAAGC--AGUCAAGCUGAAAGCC-------- ---.....(((((.(..((------((....((((((.(((........)))((..((((((.......))))))..))-......))--))))))))..))))))-------- ( -32.30, z-score = -3.13, R) >droWil1.scaffold_181089 11030242 91 + 12369635 ------CUGGCUUAUUGGC---------UGCGGCUGCGGCUAUCAAAUCAUUGGUACAACAUCAUUAACAUGUUAGCCAGAGUCUGGCUAAGUCGAGCUAAAAGCC-------- ------..(((((.(((((---------(.((((((..(.((((((....)))))))..))...........((((((((...)))))))))))))))))))))))-------- ( -31.40, z-score = -2.65, R) >droVir3.scaffold_13047 18233449 78 + 19223366 --------------UUGGC-------AUAUUGGCUGCGGCCUGGAAAUCGUCGGUACAACAUCAUUAACUUAUGGCCC--AGUCAUGU-GGGCCAAGCCUCG------------ --------------(((((-------....((((((.((((.(..(((.(..(......)..))))..)....)))))--)))))...-..)))))......------------ ( -23.30, z-score = -0.45, R) >droGri2.scaffold_15074 1285989 90 - 7742996 --------------UUGCC-------AUAUUGGUUGCGGCCAGAAAAUUGACGGUACAACGUCAUUAACUUACAACCC--AGUCAUGU-AAGCCAAACCGAACCAAACCGAGCA --------------.((((-------...((((((.(((.........(((((......)))))....((((((....--.....)))-))).....)))))))))...).))) ( -21.80, z-score = -1.88, R) >consensus ______CCGGCUUAUUGGC______UCCUUCGAUUGCGGCUAUCAAAUCAUCGGUACAACGUCAUUAACAUGCC_GCC__AAUGAAGC__AGUCAAGCCGAAAGCU________ ........(((((.(((((............(....)((((...........(((....................)))............))))..))))))))))........ ( -7.74 = -8.23 + 0.49)
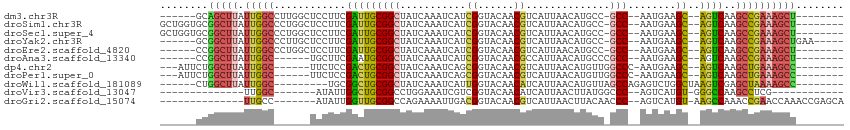

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:52:59 2011