| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,800,374 – 24,800,470 |
| Length | 96 |
| Max. P | 0.930288 |

| Location | 24,800,374 – 24,800,470 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.46 |
| Shannon entropy | 0.49431 |
| G+C content | 0.38478 |
| Mean single sequence MFE | -23.26 |
| Consensus MFE | -11.80 |
| Energy contribution | -11.90 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.930288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
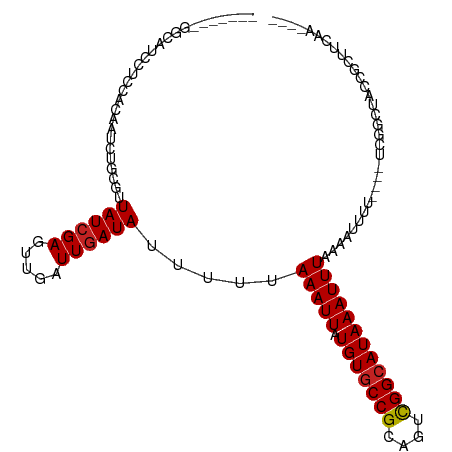
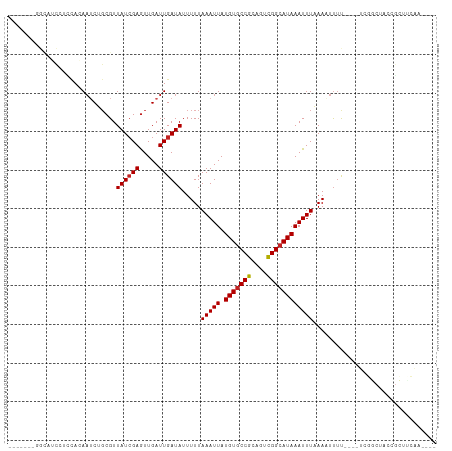
>dm3.chr3R 24800374 96 + 27905053 -------GGCAUCCUUGAGAAACGGCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUUAAAUUUU----UCGGCUACCGCUUCAA---- -------...........(((.(((((....))((((((..(((....(((((...(((((((....)))))))...))))))))..----)))))).))).)))..---- ( -22.00, z-score = -0.72, R) >droSim1.chr3R 24484261 96 + 27517382 -------GGCAUCCUUCAGAAUCGGCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUUAAAUUUU----UCGGCUACCGCUCCAA---- -------((...((.....(((((((........)))))))((.(..((((((...(((((((....)))))))...))))))..).----))))...)).......---- ( -21.90, z-score = -0.90, R) >droSec1.super_4 3662915 96 + 6179234 -------GGCAUCCUUCAGAAUCGGCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUUAAAUUUU----UCGGCUACCGCUCCAA---- -------((...((.....(((((((........)))))))((.(..((((((...(((((((....)))))))...))))))..).----))))...)).......---- ( -21.90, z-score = -0.90, R) >droEre2.scaffold_4820 7233577 96 - 10470090 -------GGCAUCCUCCAGAAUCGGCGUUAUAGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUAAAAUCUU----UCGGCUACCGCUCCAA---- -------((......))......((.((.....((((((..(((...((((((((.(((((((....))))))))))))))))))..----))))))...)).))..---- ( -24.90, z-score = -1.94, R) >droAna3.scaffold_13340 3928813 106 - 23697760 GGUAGCUGGUGGCUGCCACAAUCUGCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUAAAAUUUUCG--GCUACCGCCGCUUCCAA--- ((.(((.(((((..(((.(((((.((........)).)))))....(((((((((.(((((((....)))))))))))))))).....)--))..)))))))).))..--- ( -42.10, z-score = -5.20, R) >dp4.chr2 17557347 96 - 30794189 -----------GCUGCCACAAUCUGCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUAAAAUUUUCCUUUCGGCUACCGCUUCAA---- -----------((.(((.(((((.((........)).)))))....(((((((((.(((((((....))))))))))))))))..........)))....)).....---- ( -25.90, z-score = -3.09, R) >droPer1.super_0 7216443 96 + 11822988 -----------GCUGCCACAUUCUGCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUAAAAUUUUCCUUUCGGCUACCGCUUCAA---- -----------.............(((......((((((..((...(((((((((.(((((((....))))))))))))))))...))...))))))..))).....---- ( -23.50, z-score = -2.27, R) >droWil1.scaffold_181089 11022909 99 + 12369635 ------------GGCAUACAAUCUGCGUUAUCGAAUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUUGGCAUAAAUUUAAAAUUUUAGGCUGCCUUCUCUGAUGAUGGUG ------------(((...(((((..((....))....)))))....(((((((((.(((((((....))))))))))))))))......)))(((.((......)).))). ( -24.50, z-score = -1.80, R) >droVir3.scaffold_13047 18225900 80 + 19223366 --------GUCCUGUCCACAAGCUUCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUACCGCAGUCGGCAUAAAUUUAAAUUUGCG----------------------- --------....((((....((((.((....))))))....))))..((((((((.(((.(((....))).)))))))))))......----------------------- ( -11.40, z-score = 0.21, R) >droMoj3.scaffold_6540 20649450 81 - 34148556 -------CGUCCUACACACAAUCUGCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUAAAUUUUCC----------------------- -------...........(((((.((........)).))))).....((((((((.(((((((....)))))))))))))))......----------------------- ( -18.90, z-score = -2.78, R) >droGri2.scaffold_15074 1279268 81 - 7742996 -------CGCCUUGCACACAAUCUGCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUAAAUUUUCG----------------------- -------...........(((((.((........)).))))).....((((((((.(((((((....)))))))))))))))......----------------------- ( -18.90, z-score = -1.76, R) >consensus _______GGCAUCCUCCACAAUCUGCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUAAAAUUUU____UCGGCUACCGCUUCAA____ ............................((((((.....)))))).....(((((.(((((((....))))))))))))................................ (-11.80 = -11.90 + 0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:52:56 2011