
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,798,616 – 24,798,717 |
| Length | 101 |
| Max. P | 0.514024 |

| Location | 24,798,616 – 24,798,717 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 73.42 |
| Shannon entropy | 0.50385 |
| G+C content | 0.38671 |
| Mean single sequence MFE | -22.43 |
| Consensus MFE | -12.33 |
| Energy contribution | -12.85 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.514024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
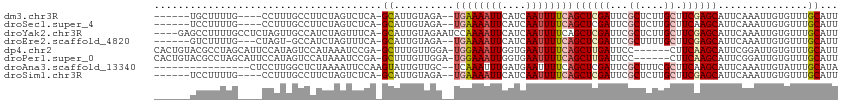
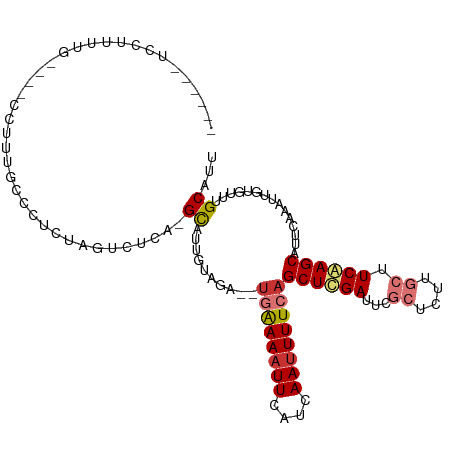
>dm3.chr3R 24798616 101 + 27905053 ------UGCUUUUG----CCUUUGCCUUCUAGUCUCA-GCAUUGUAGA--UGAAAAUUCAUCAAUUUUCAGCUCGAUUCGCUCUUGCUUCGAGCAUUCAAAUUGUGUUUGCAUU ------((((...(----.((.........)).)..)-))).((((((--((((((((....))))))).((((((...((....)).))))))...........))))))).. ( -21.50, z-score = -0.60, R) >droSec1.super_4 3661181 101 + 6179234 ------UCCUUUUG----CCUUUGCCUUCUAGUCUCA-GCAUUGUAGA--UGAAAAUUCAUCAAUUUUCAGCUCGAUUCGCUCUUGCUUCAAGCAUUCAAAUUGUGUUUGCAUU ------........----............(((...(-((((((.((.--((((((((....)))))))).))))))..)))...))).(((((((.......))))))).... ( -15.30, z-score = 0.51, R) >droYak2.chr3R 7130982 109 - 28832112 ----GAGCCUUUUGCCUCUAGUUGCCAUCUAGUUUCA-GCAUUGUAGAAUCCAAAAUUCAUCAAUUUUCAGCUCGAUUCGCUCUUGCUUCGAGCAUUCAAAUUGUGUUUGCAUU ----..((.(((((..((((..(((............-)))...))))...)))))..((.((((((...((((((...((....)).))))))....))))))))...))... ( -19.60, z-score = -0.10, R) >droEre2.scaffold_4820 7231865 101 - 10470090 ------GUCUUUUG---CUAGU-GCCAUCUAGUUUCA-GCAUUGUAGA--UGAAAAUUCAUCAAUUUUCAGCUCGAUUCGCUUUUGCUUCGAGCAUUCAAAUUGUGUUUGCAUU ------(.((...(---((((.-.....)))))...)-))..((((((--((((((((....))))))).((((((...((....)).))))))...........))))))).. ( -24.80, z-score = -1.92, R) >dp4.chr2 17555400 106 - 30794189 CACUGUACGCCUAGCAUUCCAUAGUCCAUAAAUCCGA-GCUUUGUUGGA-UGGAAAUUGGUGAAUUUUCAGCUUGAUUCC------CUUCAAGCAUUCGGAUUGUGUUUGCAUU .............(((...((((((((....((((((-......)))))-)(((((((....))))))).((((((....------..))))))....))))))))..)))... ( -31.00, z-score = -2.56, R) >droPer1.super_0 7214476 106 + 11822988 CACUGUACGCCUAGCAUUCCAUAGUCCAUAAAUCCGA-GCUUUGUUGGA-UGGAAAUUGGUGAAUUUUCAGCUUGAUUCC------CUUCAAGCAUUCGGAUUGUGUUUGCAUU .............(((...((((((((....((((((-......)))))-)(((((((....))))))).((((((....------..))))))....))))))))..)))... ( -31.00, z-score = -2.56, R) >droAna3.scaffold_13340 3927099 96 - 23697760 ----------------CUCCUUGGCUCUAAAAUUCCAAGUAUUGUUGC--UCAAAUUUGAUGAAUUUUCAGCUCGAUUCGCUUUCGCUUCAAGCAUUCAAAUUGUAUUUGCAUA ----------------...(((((..........)))))......(((--...((((((((((....)))(((.((..((....))..)).)))..)))))))......))).. ( -15.30, z-score = -0.04, R) >droSim1.chr3R 24482542 101 + 27517382 ------UCCUUUUG----CCUUUGCCUUCUAGUCUCA-GCAUUGUAGA--UGAAAAUUCAUCAAUUUUCAGCUCGAUUCGCUCUUGCUUCGAGCAUUCAAAUUGUGUUUGCAUU ------......((----(....((......))....-))).((((((--((((((((....))))))).((((((...((....)).))))))...........))))))).. ( -20.90, z-score = -1.14, R) >consensus ______UCCUUUUG____CCUUUGCCCUCUAGUCUCA_GCAUUGUAGA__UGAAAAUUCAUCAAUUUUCAGCUCGAUUCGCUCUUGCUUCAAGCAUUCAAAUUGUGUUUGCAUU ......................................((..........((((((((....))))))))((((((...((....)).))))))...............))... (-12.33 = -12.85 + 0.52)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:52:55 2011