
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,783,762 – 24,783,861 |
| Length | 99 |
| Max. P | 0.564145 |

| Location | 24,783,762 – 24,783,861 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 66.01 |
| Shannon entropy | 0.67520 |
| G+C content | 0.52966 |
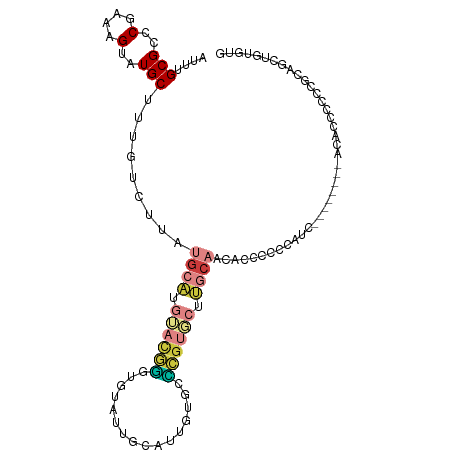
| Mean single sequence MFE | -26.61 |
| Consensus MFE | -9.80 |
| Energy contribution | -10.60 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.564145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
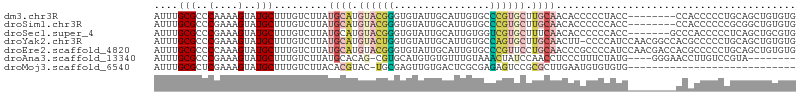
>dm3.chr3R 24783762 99 - 27905053 AUUUGCGCCCAAAAGUAUGCUUUGUCUUAUGCAUGUACGGGUGUAUUGCAUUGUGCCCGUGCUUGCAACACCCCCUACC--------CCACCCCCUGCAGCUGUGUG ..((((.....((((....))))......((((.(((((((..((......))..))))))).))))............--------.........))))....... ( -26.40, z-score = -1.34, R) >droSim1.chr3R 24467483 99 - 27517382 AUUUGCGCCCGAAAGUAUGCUUUGUCUUAUGCAUGUACGGGUGUAUUGCAUUGUGCCCGUGCUUGCAACACCCCCCACC--------CCACCCCCCGCGGCUGUGUG ....(((..(....)..))).........((((.(((((((..((......))..))))))).))))((((..(((...--------.........).))..)))). ( -30.40, z-score = -1.87, R) >droSec1.super_4 3646452 100 - 6179234 AUUUGCGCCCGAAAGUAUGCUUUGUCUUAUGCAUGUACGGGUGUAUUGCAUUGUGGUCGUGCUUUCAACACCCCCCACC-------GCCCACCCCCUCAGCUGCGUG ...((((((((....(((((..........)))))..))))))))..((((((.((..(((..................-------...)))..)).))).)))... ( -24.60, z-score = -0.38, R) >droYak2.chr3R 7115771 106 + 28832112 AUUUGCGCCCGAAAGUAUGCUUUGUCUUAUGCAUGUACUGGUGUAUUGCAUUGUGCCAGUGCUUGCAACUU-CCCCAUCCAACGGCCACGCCCCCUGCAGCUGUGUG ....(((..(....)..))).........((((.(((((((..((......))..))))))).))))....-.........(((((...((.....)).)))))... ( -31.00, z-score = -1.41, R) >droEre2.scaffold_4820 7217207 107 + 10470090 AUUUGCGCCCCAAAGUAUGCUUUGUCUUAUGCAUGUACGGGUGUAUUGCAUUGUGCCCGUUCCUGCAACCCGCCCCAUCCAACGACCACGCCCCCUGCAGCUGUGUG ....(((...(((((....))))).....((((.(.(((((..((......))..))))).).))))...))).............(((((...........))))) ( -26.30, z-score = -0.53, R) >droAna3.scaffold_13340 3911479 94 + 23697760 AUUUGCGCCCGAAAGUAUGCUUUGUCUUAUGCACAG-CGUGCAUGUGUGUUUGUAAACUAUCCAACCUCCCUUUCUAUG----GGGAACCUUGUCCGUA-------- ...((((..(((..(((((((.(((.....))).))-))))).........................(((((......)----))))...)))..))))-------- ( -22.00, z-score = -1.21, R) >droMoj3.scaffold_6540 20631730 78 + 34148556 AUUUGCGCUCGAAAGUAUGCUUUGUCUUACACGUAC-UGCGAGUUGUGACUCGCGAGAGUCCGCGCUUGAAUGUGUGUG---------------------------- ....(((..(....)..))).......(((((((..-..(((((.(((((((....)))).)))))))).)))))))..---------------------------- ( -25.60, z-score = -1.63, R) >consensus AUUUGCGCCCGAAAGUAUGCUUUGUCUUAUGCAUGUACGGGUGUAUUGCAUUGUGCCCGUGCUUGCAACACCCCCCAUC________ACACCCCCCGCAGCUGUGUG ....(((..(....)..))).........((((.((((((................)))))).))))........................................ ( -9.80 = -10.60 + 0.80)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:52:52 2011