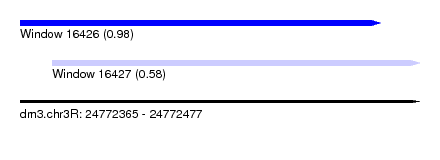
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,772,365 – 24,772,477 |
| Length | 112 |
| Max. P | 0.983364 |

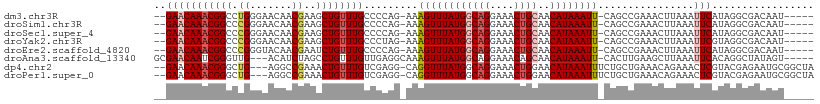
| Location | 24,772,365 – 24,772,466 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.47 |
| Shannon entropy | 0.36157 |
| G+C content | 0.44452 |
| Mean single sequence MFE | -28.79 |
| Consensus MFE | -16.92 |
| Energy contribution | -17.73 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.983364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
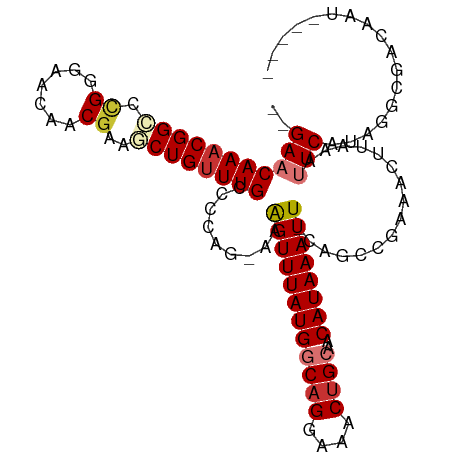
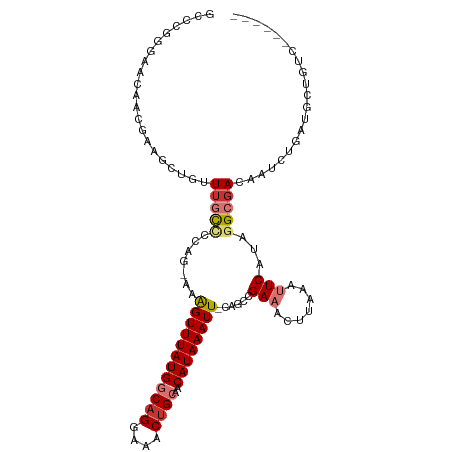
>dm3.chr3R 24772365 101 + 27905053 --GAACAAACGGCCUGGGAACAACGAAGCUGUUUGCCCCAG-AAAGUUUAUGGCAGGAAACUGCAACAUAAAUU-CAGCCGAAACUUAAAUUCAUAGGCGACAAU----- --.......(((((((((...((((....))))...)))))-..((((((((((((....))))..))))))))-..))))...............(....)...----- ( -29.00, z-score = -3.28, R) >droSim1.chr3R 24460610 101 + 27517382 --GAACAAACGGCCCGGGAACAACGAAGCUGUUUGCCCCAG-AAAGUUUAUGGCAGGAAACUGCAACAUAAAUU-CAGCCGAAACUUAAAUUCAUAGGCGACAAU----- --...((((((((.((.......))..))))))))......-..((((((((((((....))))..))))))))-..(((................)))......----- ( -27.09, z-score = -2.62, R) >droSec1.super_4 3635261 101 + 6179234 --GAACAAACGGCCCGGGAACAACGAAGCUGUUUGCCCCAG-AAAGUUUAUGGCAGGAAACUGCAACAUAAAUU-CAGCCGAAACUUAAAUUCAUAGGCGACAAU----- --...((((((((.((.......))..))))))))......-..((((((((((((....))))..))))))))-..(((................)))......----- ( -27.09, z-score = -2.62, R) >droYak2.chr3R 7103673 101 - 28832112 --GAACAAACGGCCCGGGAACAACGAAGCUGUUUGCCCUAG-AAAGUUUAUGGCAGGAAACUGCAACAUAAAUU-CAGCCGAAACUUAAAUUCGUAGGCGACAAU----- --...((((((((.((.......))..))))))))(((((.-..((((((((((((....))))..))))))))-....((((.......)))))))).).....----- ( -27.70, z-score = -2.50, R) >droEre2.scaffold_4820 7205698 101 - 10470090 --GAACAAACGGCCCGGGUACAACGAAUCUGUUUGCCCCAG-AAAGUUUAUGGCAGGAAACUGCAACAUAAAUU-CAGCCGAAACUUAAAUUCAUAGGCGACAAU----- --.......((((..(((((.((((....)))))))))...-..((((((((((((....))))..))))))))-..))))...............(....)...----- ( -27.80, z-score = -2.90, R) >droAna3.scaffold_13340 3900039 101 - 23697760 GCGAACAAUCGGGUUG---ACAUCUAGCCUGUUUGUUGAGGCAAAGUUUAUGGCAGGAAACAGCAACAUAAAUU-CACUUGAAGCUUAAAUUCACAGGCUAUAGU----- .(((....))).....---.....((((((((..(((.((((....((((((((.(....).))..))))))((-(....))))))).)))..))))))))....----- ( -25.80, z-score = -1.65, R) >dp4.chr2 17525207 104 - 30794189 --GAACAAACGGGCUG---AGGCCGAAACUGUUUGUCGAGG-CAGGUUUAUGGCAGGAAACUGGAACAUAAAUUUCUGCUGAAACAGAAACUCGUACGAGAAUGCGGCUA --..............---.(((((...((((((.....((-((((((((((.(((....)))...)))))))..))))).))))))...(((....)))....))))). ( -32.90, z-score = -3.18, R) >droPer1.super_0 7183702 104 + 11822988 --GAACAAACGGGCUG---AGGCCGAAACUGUUUGUCGAGG-CAGGUUUAUGGCAGGAAACUGGAACAUAAAUUUCUGCUGAAACAGAAACUCGUACGAGAAUGCGGCUA --..............---.(((((...((((((.....((-((((((((((.(((....)))...)))))))..))))).))))))...(((....)))....))))). ( -32.90, z-score = -3.18, R) >consensus __GAACAAACGGCCCGGGAACAACGAAGCUGUUUGCCCCAG_AAAGUUUAUGGCAGGAAACUGCAACAUAAAUU_CAGCCGAAACUUAAAUUCAUAGGCGACAAU_____ ..(((((((((((..............)))))))).........((((((((((((....))))..))))))))................)))................. (-16.92 = -17.73 + 0.81)
| Location | 24,772,374 – 24,772,477 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.69 |
| Shannon entropy | 0.41831 |
| G+C content | 0.45615 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -12.46 |
| Energy contribution | -13.21 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.580044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24772374 103 + 27905053 GCCUGGGAACAACGAAGCUGUUUGCCCCAG-AAAGUUUAUGGCAGGAAACUGCAACAUAAAUU-CAGCCGAAACUUAAAUUCAUAGGCGACAAUCUGAUGCUGUC------ ..(((((...((((....))))...)))))-..((((((((((((....))))..))))))))-..(((................)))((((.........))))------ ( -26.59, z-score = -1.40, R) >droSim1.chr3R 24460619 103 + 27517382 GCCCGGGAACAACGAAGCUGUUUGCCCCAG-AAAGUUUAUGGCAGGAAACUGCAACAUAAAUU-CAGCCGAAACUUAAAUUCAUAGGCGACAAUCUGAUGCUGUC------ ((..(((...((((....))))..)))(((-(.((((((((((((....))))..))))))))-..(((................))).....))))..))....------ ( -26.59, z-score = -1.43, R) >droSec1.super_4 3635270 103 + 6179234 GCCCGGGAACAACGAAGCUGUUUGCCCCAG-AAAGUUUAUGGCAGGAAACUGCAACAUAAAUU-CAGCCGAAACUUAAAUUCAUAGGCGACAAUCUGAUGCUGUC------ ((..(((...((((....))))..)))(((-(.((((((((((((....))))..))))))))-..(((................))).....))))..))....------ ( -26.59, z-score = -1.43, R) >droYak2.chr3R 7103682 103 - 28832112 GCCCGGGAACAACGAAGCUGUUUGCCCUAG-AAAGUUUAUGGCAGGAAACUGCAACAUAAAUU-CAGCCGAAACUUAAAUUCGUAGGCGACAAUCUGAUGCUGUC------ ((..(((...((((....))))..)))(((-(.((((((((((((....))))..))))))))-..(((...((........)).))).....))))..))....------ ( -25.40, z-score = -0.87, R) >droEre2.scaffold_4820 7205707 103 - 10470090 GCCCGGGUACAACGAAUCUGUUUGCCCCAG-AAAGUUUAUGGCAGGAAACUGCAACAUAAAUU-CAGCCGAAACUUAAAUUCAUAGGCGACAAUCUGAUGCUGUC------ ((..(((((.((((....)))))))))(((-(.((((((((((((....))))..))))))))-..(((................))).....))))..))....------ ( -29.09, z-score = -2.38, R) >droAna3.scaffold_13340 3900050 101 - 23697760 ---GGUUGACAUCUAGCCUGUUUGUUGAGGCAAAGUUUAUGGCAGGAAACAGCAACAUAAAUU-CACUUGAAGCUUAAAUUCACAGGCUAUAGUCGGAGGAUGUU------ ---..(((((...((((((((..(((.((((....((((((((.(....).))..))))))((-(....))))))).)))..))))))))..)))))........------ ( -28.60, z-score = -2.20, R) >dp4.chr2 17525216 106 - 30794189 ---GGCUGAGGCCGAAACUGUUUGUCGAGG-CAGGUUUAUGGCAGGAAACUGGAACAUAAAUUUCUGCUGAAACAGAAACUCGUACGAGA-AUGCGGCUACCGCCACUGUU ---(((.(.(((((...((((((.....((-((((((((((.(((....)))...)))))))..))))).))))))...(((....))).-...))))).).)))...... ( -39.30, z-score = -3.82, R) >droPer1.super_0 7183711 106 + 11822988 ---GGCUGAGGCCGAAACUGUUUGUCGAGG-CAGGUUUAUGGCAGGAAACUGGAACAUAAAUUUCUGCUGAAACAGAAACUCGUACGAGA-AUGCGGCUACCGCCACUGUU ---(((.(.(((((...((((((.....((-((((((((((.(((....)))...)))))))..))))).))))))...(((....))).-...))))).).)))...... ( -39.30, z-score = -3.82, R) >consensus GCCCGGGAACAACGAAGCUGUUUGCCCCAG_AAAGUUUAUGGCAGGAAACUGCAACAUAAAUU_CAGCCGAAACUUAAAUUCAUAGGCGACAAUCUGAUGCUGUC______ .....................(((((.......((((((((((((....))))..))))))))......(((.......)))...)))))..................... (-12.46 = -13.21 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:52:50 2011