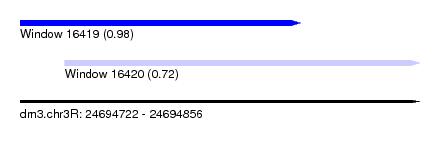
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,694,722 – 24,694,856 |
| Length | 134 |
| Max. P | 0.980052 |

| Location | 24,694,722 – 24,694,816 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 76.25 |
| Shannon entropy | 0.50592 |
| G+C content | 0.53056 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -18.61 |
| Energy contribution | -18.62 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24694722 94 + 27905053 ------------AUGCCGGCAUCUGCA---AUAGCCUCAUCCACAUCUAGUCAUCAGCACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCG ------------.(((.(((.......---...)))......((.....)).....))).(((((((((((...(((.....)))...)).)))))))))......... ( -22.30, z-score = -0.96, R) >droSim1.chr3R 24377737 94 + 27517382 ------------AUGCCGGCAUCUGCA---AUAGCCUCAUCCUCAUCUAGUCAUCAGCACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCG ------------.....(((.......---...)))........................(((((((((((...(((.....)))...)).)))))))))......... ( -22.10, z-score = -0.93, R) >droSec1.super_4 3558072 94 + 6179234 ------------AUGCCGGCAUCUGCA---AUAGCCUCAUCCUCAUCUAGUCAUCAGCACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCG ------------.....(((.......---...)))........................(((((((((((...(((.....)))...)).)))))))))......... ( -22.10, z-score = -0.93, R) >droYak2.chr3R 7022143 94 - 28832112 ------------CUACCGGCAUCUGCA---AUCGCCACAUCUUCAUCUAGUCAUCAGCACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCG ------------.....(((.......---...)))........................(((((((((((...(((.....)))...)).)))))))))......... ( -22.60, z-score = -2.00, R) >droEre2.scaffold_4820 7127218 94 - 10470090 ------------CUGCCAGCAUCUGCA---AUAGCCUCAUCUUCAUCUAGUCAUCAGCACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCG ------------......((....)).---...((.....((......))......))..(((((((((((...(((.....)))...)).)))))))))......... ( -20.80, z-score = -0.95, R) >droAna3.scaffold_13340 15806551 91 - 23697760 ------------------UCAUCCACAACCGCCGUUGCAUCUUCAUCUAGUCACCAGCACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCG ------------------...............((((...((......))....))))..(((((((((((...(((.....)))...)).)))))))))......... ( -21.60, z-score = -2.42, R) >dp4.chr2 16805885 105 - 30794189 ----UCACAGCCCCACCGGCAUCCGCAUUGACAUCUUCAUUGGCAUCUAGUCACCAGCACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCG ----.....(((.....)))....((..(((.....))).((((.....))))...))..(((((((((((...(((.....)))...)).)))))))))......... ( -27.10, z-score = -1.83, R) >droPer1.super_0 6453770 105 + 11822988 ----UCACAGCCCCACCGGCAUCCGCAUUGACAUCCUCAUUGGCAUCUAGUCACCAGCACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCG ----.....(((.....)))....((..((((((((.....)).))...))))...))..(((((((((((...(((.....)))...)).)))))))))......... ( -27.60, z-score = -1.94, R) >droWil1.scaffold_181130 9485459 89 - 16660200 --------------------GGCGAUUGCAGCGACAACAUCUUCAUCGAAUCACCCACACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAGCAUCCACCCG --------------------.((....(((((((((.......(((((...........)))))(..((.(.....).))..)....)))))))))..))......... ( -24.30, z-score = -1.98, R) >droMoj3.scaffold_6540 24975827 100 - 34148556 ------GGGCCUGCGCCGGUGCCGGUU---GCUGUUGAGGCUGCCGUCAGUCAUCCACCUGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCACCCACCCG ------.(((....)))((((.((((.---(((((((..(((((((((((........)))))))))))((((...))))..))))).)).)))).))))......... ( -37.20, z-score = -1.00, R) >droVir3.scaffold_13047 10164625 97 + 19223366 ------------GCGUCCACGGCUGUGGAGGCGGCCACAGCUGCAGCCAGUCAUCCACACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCG ------------.......(((.((((((((((((..(....)..))).))).)))))).(((((((((((...(((.....)))...)).)))))))))......))) ( -36.20, z-score = -1.88, R) >droGri2.scaffold_14906 6868237 109 - 14172833 CCGCCGCCAGCUGUCAAUUCUGCUGUUGAAGCAGCCACAUCUUCAUCUAGUCAUCAUCAUGAUGGCAGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCG .........(((((((((......))))..))))).......................(((((((((((((...(((.....)))...)).)))))))))))....... ( -28.30, z-score = -2.10, R) >consensus ____________CUGCCGGCAUCUGCA___ACAGCCUCAUCUUCAUCUAGUCAUCAGCACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCG ............................................................(((((((((((...(((.....)))...)).)))))))))......... (-18.61 = -18.62 + 0.01)
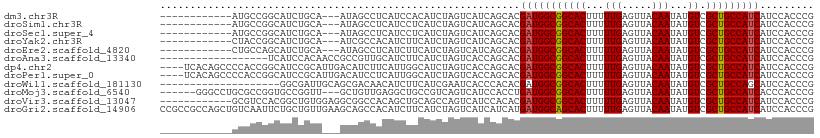
| Location | 24,694,737 – 24,694,856 |
|---|---|
| Length | 119 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.28 |
| Shannon entropy | 0.26386 |
| G+C content | 0.51855 |
| Mean single sequence MFE | -23.61 |
| Consensus MFE | -18.61 |
| Energy contribution | -18.62 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.719468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24694737 119 + 27905053 -AUAGCCUCAUCCACAUCUAGUCAUCAGCACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCGAUCAGCCACAUCAGCCACACAAGCCAUCCUCAUCCACGUC -...((..............((((((.....))))))(((((((...))))......((((.((((..(((........))))))).))))..)))......))................ ( -21.30, z-score = -1.10, R) >droSim1.chr3R 24377752 119 + 27517382 -AUAGCCUCAUCCUCAUCUAGUCAUCAGCACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCGAUCAGCCACAUCAGCCACACAAGCCAUCCUCAUCCACGUC -...((..............((((((.....))))))(((((((...))))......((((.((((..(((........))))))).))))..)))......))................ ( -21.30, z-score = -1.05, R) >droSec1.super_4 3558087 119 + 6179234 -AUAGCCUCAUCCUCAUCUAGUCAUCAGCACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCGAUCAGCCACAUCAGCCACACAAGCCAUCCUCAUCCACGUC -...((..............((((((.....))))))(((((((...))))......((((.((((..(((........))))))).))))..)))......))................ ( -21.30, z-score = -1.05, R) >droYak2.chr3R 7022158 119 - 28832112 -AUCGCCACAUCUUCAUCUAGUCAUCAGCACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCGAUCAGCCACAUCAGCCACACAAGCCAUCCUCAUCCACGUC -...((..............((((((.....))))))(((((((...))))......((((.((((..(((........))))))).))))..)))......))................ ( -21.40, z-score = -1.09, R) >droEre2.scaffold_4820 7127233 119 - 10470090 -AUAGCCUCAUCUUCAUCUAGUCAUCAGCACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCGAUCAGCCACAUCAGCCACACAAGCCAUCCUCAUCCACGUC -...((..............((((((.....))))))(((((((...))))......((((.((((..(((........))))))).))))..)))......))................ ( -21.30, z-score = -1.08, R) >droAna3.scaffold_13340 15806566 116 - 23697760 ----GUUGCAUCUUCAUCUAGUCACCAGCACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCGAUCAGCCUUACCAGCCACACAAGCCAUCCUCAUCCACAUC ----((((.(((........((.....))..(((((((((((...(((.....)))...)).)))))))))........))))))).................................. ( -22.90, z-score = -1.69, R) >dp4.chr2 16805910 120 - 30794189 GACAUCUUCAUUGGCAUCUAGUCACCAGCACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCGAUCAGCCACAUCAGCCACACAAGCCAUCCUCUUCCACAUC ...........((((................(((((((((((...(((.....)))...)).)))))))))........(((.......)))..........)))).............. ( -25.00, z-score = -1.53, R) >droPer1.super_0 6453795 120 + 11822988 GACAUCCUCAUUGGCAUCUAGUCACCAGCACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCGAUCAGCCACAUCAGCCACAUAAGCCAUCCUCUUCCACAUC ...........((((................(((((((((((...(((.....)))...)).)))))))))........(((.......)))..........)))).............. ( -25.00, z-score = -1.49, R) >droWil1.scaffold_181130 9485469 119 - 16660200 -GCGACAACAUCUUCAUCGAAUCACCCACACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAGCAUCCACCCGAUCAGCCGCAUCAGCCACACAAGCCAUCCUCGUCGACCUC -.((((....((......))...........(((((((((((((...))))......((((.((((.....(((.....))))))).))))..)))......))))))...))))..... ( -23.30, z-score = -0.42, R) >droMoj3.scaffold_6540 24975857 110 - 34148556 ----------GCUGCCGUCAGUCAUCCACCUGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCACCCACCCGAUCAGCCGCAUCAGCCACACAAGCCAUCCUCGUCCACCUC ----------(((((((((((........))))))))))).......(((.......((((.((((..(((........))))))).))))..((.......))....)))......... ( -30.20, z-score = -3.08, R) >droVir3.scaffold_13047 10164652 110 + 19223366 ----------GCUGCAGCCAGUCAUCCACACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCGAUCAGCCGCAUCAACCACACAAACCAUCCUCGUCCACAUC ----------(.(((.((..(((........(((((((((((...(((.....)))...)).)))))))))........)))..)).))).)............................ ( -25.29, z-score = -2.35, R) >droGri2.scaffold_14906 6868267 119 - 14172833 -GCAGCCACAUCUUCAUCUAGUCAUCAUCAUGAUGGCAGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCGAUCAGCCGCAUCAGCCACACAAACCAUCCUCAUCGACAUC -...................(((......(((((((((((((...(((.....)))...)).)))))))))))..............((....))...................)))... ( -25.00, z-score = -2.74, R) >consensus _AUAGCCUCAUCUUCAUCUAGUCAUCAGCACGAUGGCGGCACUUUUUGAGUUACAAUAUGUCGCUGCCAUCAUCCACCCGAUCAGCCACAUCAGCCACACAAGCCAUCCUCAUCCACAUC ...............................(((((((((((...(((.....)))...)).)))))))))................................................. (-18.61 = -18.62 + 0.01)

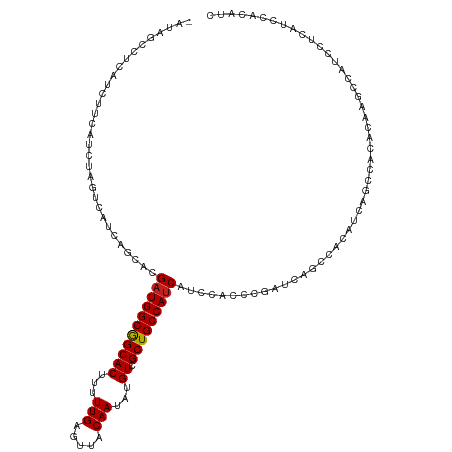
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:52:44 2011