| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,683,435 – 24,683,529 |
| Length | 94 |
| Max. P | 0.998902 |

| Location | 24,683,435 – 24,683,529 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 68.06 |
| Shannon entropy | 0.55802 |
| G+C content | 0.51891 |
| Mean single sequence MFE | -31.74 |
| Consensus MFE | -16.48 |
| Energy contribution | -19.00 |
| Covariance contribution | 2.52 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.54 |
| SVM RNA-class probability | 0.998902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
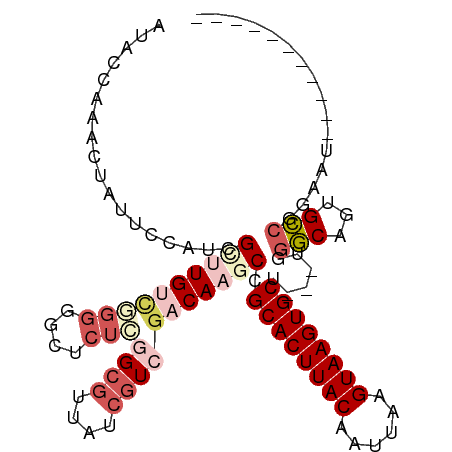
>dm3.chr3R 24683435 94 + 27905053 AUACCGAACCAUUCCAUCGGUUGUCUGGGUCUCUCGGCGUUAUCGUCGACAAGCCGCACUUACAAUUAGGUAAGUGCU---UGGGCAGUGCCGGAAU----------- ...(((((((........))))((((.((..(.((((((....)))))).)..))((((((((......)))))))).---.)))).....)))...----------- ( -33.90, z-score = -2.55, R) >droSim1.chr3R 24366452 94 + 27517382 AUACCAUACUAUUCCAUCGCUUGUCGGGGGCUCUCGGCGUUAUCGUCGACAAGCCGCACUUACAAUUAAGUAAGUGCU---UGGGCAGUGCCGGAAU----------- ..........(((((..((((.(((.((((((.((((((....))))))..))))((((((((......)))))))))---).)))))))..)))))----------- ( -37.20, z-score = -3.92, R) >droSec1.super_4 3546727 94 + 6179234 AUACCAAACUAUUCCAUCGCUUGUCGGGGGCUCUCGGCGUUAUCGUCGACAAGCCGCACUUACAAUUAAGUAAGUGCU---UGGGCAGUGCCGGAAU----------- ..........(((((..((((.(((.((((((.((((((....))))))..))))((((((((......)))))))))---).)))))))..)))))----------- ( -37.20, z-score = -4.00, R) >droAna3.scaffold_13340 15794959 83 - 23697760 --------------CCUCGUCUGGUCGGAAUUCUGUGCGAUAUCGUCGACAAGCCGCACUUACAAUUAAGUAAGUGCUGCCUGGGCAGUGCCGGAAC----------- --------------((....(((.((((.....(((.((((...))))))).((.((((((((......)))))))).)))))).)))....))...----------- ( -27.40, z-score = -1.48, R) >droMoj3.scaffold_6540 24961876 102 - 34148556 ACAGCACACAUUUCCGUUGGCUGC------CUUAUCGCUCUGCCCUCUACAAGCCGCACUUACAAUUAAGUAAGUGCCUCUCCCUCAGAGAUUGCUCUGUCCCUCUCU ((((..((.(((((.(..(((.((------......))...)))..)........((((((((......))))))))..........)))))))..))))........ ( -23.00, z-score = -2.33, R) >consensus AUACCAAACUAUUCCAUCGCUUGUCGGGGGCUCUCGGCGUUAUCGUCGACAAGCCGCACUUACAAUUAAGUAAGUGCU___UGGGCAGUGCCGGAAU___________ ..................((((((((((....)))((((....))))))))))).((((((((......))))))))......(((...)))................ (-16.48 = -19.00 + 2.52)


| Location | 24,683,435 – 24,683,529 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 68.06 |
| Shannon entropy | 0.55802 |
| G+C content | 0.51891 |
| Mean single sequence MFE | -29.94 |
| Consensus MFE | -16.98 |
| Energy contribution | -17.22 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.996058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
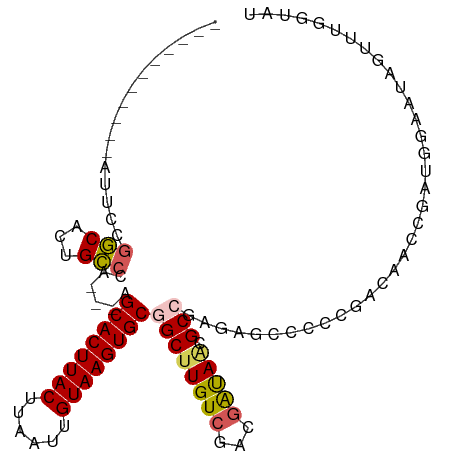
>dm3.chr3R 24683435 94 - 27905053 -----------AUUCCGGCACUGCCCA---AGCACUUACCUAAUUGUAAGUGCGGCUUGUCGACGAUAACGCCGAGAGACCCAGACAACCGAUGGAAUGGUUCGGUAU -----------....(((((..(((..---.((((((((......))))))))))).)))))........((((((....(((.........))).....)))))).. ( -30.20, z-score = -2.24, R) >droSim1.chr3R 24366452 94 - 27517382 -----------AUUCCGGCACUGCCCA---AGCACUUACUUAAUUGUAAGUGCGGCUUGUCGACGAUAACGCCGAGAGCCCCCGACAAGCGAUGGAAUAGUAUGGUAU -----------...(((..((((.(((---.((((((((......)))))))).((((((((.((....)).(....)....))))))))..)))..)))).)))... ( -31.70, z-score = -2.86, R) >droSec1.super_4 3546727 94 - 6179234 -----------AUUCCGGCACUGCCCA---AGCACUUACUUAAUUGUAAGUGCGGCUUGUCGACGAUAACGCCGAGAGCCCCCGACAAGCGAUGGAAUAGUUUGGUAU -----------...((((.((((.(((---.((((((((......)))))))).((((((((.((....)).(....)....))))))))..)))..))))))))... ( -31.80, z-score = -2.88, R) >droAna3.scaffold_13340 15794959 83 + 23697760 -----------GUUCCGGCACUGCCCAGGCAGCACUUACUUAAUUGUAAGUGCGGCUUGUCGACGAUAUCGCACAGAAUUCCGACCAGACGAGG-------------- -----------....(((..(((..(((((.((((((((......)))))))).)))))..(.((....))).)))....)))...........-------------- ( -24.60, z-score = -1.37, R) >droMoj3.scaffold_6540 24961876 102 + 34148556 AGAGAGGGACAGAGCAAUCUCUGAGGGAGAGGCACUUACUUAAUUGUAAGUGCGGCUUGUAGAGGGCAGAGCGAUAAG------GCAGCCAACGGAAAUGUGUGCUGU ........((((.(((..(((((...(((..((((((((......))))))))..))).)))))(((...((......------)).)))..........))).)))) ( -31.40, z-score = -1.78, R) >consensus ___________AUUCCGGCACUGCCCA___AGCACUUACUUAAUUGUAAGUGCGGCUUGUCGACGAUAACGCCGAGAGCCCCCGACAACCGAUGGAAUAGUUUGGUAU ................(((...)))......((((((((......))))))))((((((((...))))).)))................................... (-16.98 = -17.22 + 0.24)

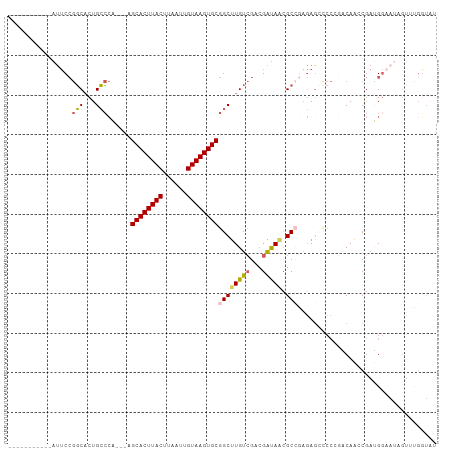
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:52:42 2011