| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,672,541 – 24,672,632 |
| Length | 91 |
| Max. P | 0.564851 |

| Location | 24,672,541 – 24,672,632 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 62.18 |
| Shannon entropy | 0.71800 |
| G+C content | 0.37898 |
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -6.37 |
| Energy contribution | -5.97 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.79 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.564851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
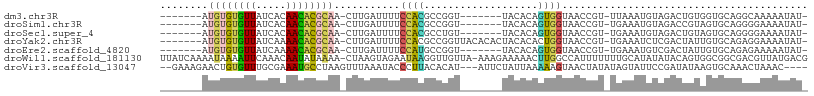
>dm3.chr3R 24672541 91 - 27905053 -------AUGUGUGUUAUCACAACACGCAA-CUUGAUUUUCCACGCCGGU-------UACACAGUGGUAACCGU-UUAAAUGUAGACUGUGGUGCAGGCAAAAAUAU- -------.((((((((.....)))))))).-((((.....((((...(((-------(((......))))))((-(((....))))).))))..)))).........- ( -26.70, z-score = -1.62, R) >droSim1.chr3R 24355683 91 - 27517382 -------AUGUGUGUUAUCACAACACGCAA-CUUGAUUUUCCACGCCGGU-------UACACAGUGGUAACCGU-UGAAAUGUAGACCGUAGUGCAGGGGAAAAUAU- -------.((((((((.....)))))))).-....(((((((.((.((((-------(((......))))))).-)).........((........)))))))))..- ( -27.80, z-score = -1.85, R) >droSec1.super_4 3536004 91 - 6179234 -------AUGUGUGUUAUCACAACACGCAA-CUUGAUUUUCCACGCCUGU-------UACACAGUGGUAACCGU-UGAAAUGUAGACUGUAGUGCAGGGGAAAAUAU- -------.((((((((.....)))))))).-....(((((((...(((((-------.(((((((.........-..........))))).))))))))))))))..- ( -25.41, z-score = -1.26, R) >droYak2.chr3R 6999581 98 + 28832112 -------AUGUGUGUUAUCAAAACACGCAA-CUUGAUUUUCCACGCCGGUUACACACUACACACUGGUAACCGU-UGAAAUCUCGACUAUUGUGCAGAGGAAAAUAU- -------.((((((((.....)))))))).-....(((((((.((((((((((.((........))))))))((-(((....)))))....).)).).)))))))..- ( -29.00, z-score = -3.41, R) >droEre2.scaffold_4820 7105114 91 + 10470090 -------AUGUGUGUUAUCAAAACACGCAA-CUUGAUUUUCCAUGCCGGU-------UACACAGUGGUAACCGU-UGAAAUGUCGACUAUUGUGCAGAGAAAAAUAU- -------.((((((((.....)))))))).-.....(((((..(((((((-------(((......))))))((-(((....)))))....).)))..)))))....- ( -27.40, z-score = -3.04, R) >droWil1.scaffold_181130 9460231 106 + 16660200 UUAUCAAAAUAAAAUUCAAACAAUAUAAAA-CUAAGUAGAAUAAGGUUGUUA-AAAGAAAAACUUGGCCAUUUUUUUGCAUAUAUACAGUGGCGGCGACGUUAUGACG ..............................-....((...((((.((((((.-.(((.....))).((((((...............)))))))))))).)))).)). ( -14.96, z-score = -0.16, R) >droVir3.scaffold_13047 10137456 99 - 19223366 --GAAAGAACUGUGUUUGCGAAAUGCCUAAGUUUAAAUACCCUUACACAU---AUUCUAUUAAAAAGUAACUAUAUAGUAUUCCGAUAUAAGUGCAAACUAAAC---- --...........((((((((((((..((((..........))))..)))---.)))............(((.((((.........))))))))))))).....---- ( -10.40, z-score = 0.11, R) >consensus _______AUGUGUGUUAUCACAACACGCAA_CUUGAUUUUCCACGCCGGU_______UACACAGUGGUAACCGU_UGAAAUGUAGACUGUAGUGCAGAGGAAAAUAU_ ........((((((((.....))))))))...........((((....((........))...))))......................................... ( -6.37 = -5.97 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:52:40 2011