| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,656,063 – 24,656,157 |
| Length | 94 |
| Max. P | 0.742496 |

| Location | 24,656,063 – 24,656,157 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 66.57 |
| Shannon entropy | 0.64827 |
| G+C content | 0.52565 |
| Mean single sequence MFE | -23.41 |
| Consensus MFE | -10.55 |
| Energy contribution | -11.03 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24656063 94 - 27905053 UUUGUGUUCCUCC------GCCUUCGCCGAGCAAAUCCCACGGCUUGAUGAAGCACUUCCUGCGGCUCAAGUGUCGCCCA----UCGUGUGCGAAUUUCACUCG ...(((......(------(((((((.(((((..........))))).)))))(((.....(((((......)))))...----..))).))).....)))... ( -23.60, z-score = -0.41, R) >droSim1.chr3R 24337338 100 - 27517382 UUUGUGUUCCUCCGCCAACGCCUUCACCGAGCAAAUCCCACAGCUUGAUGAAGCACUUCCUGCGGCUCAAGUGUCGCCCA----UCGUGUGCGAAUUUCACUCG ...(((......(((..(((((((((.(((((..........))))).)))))........(((((......)))))...----..))))))).....)))... ( -25.70, z-score = -1.57, R) >droSec1.super_4 3519504 100 - 6179234 UUUGUGUUCCUCCGCCAACGCCUUCACCGAGCAAAUCCCACAGCUUGAUGAAGCACUUCCUGCGGCUCAAGUGUCGCCCA----UCGUGUGCGAAUUUCACUCG ...(((......(((..(((((((((.(((((..........))))).)))))........(((((......)))))...----..))))))).....)))... ( -25.70, z-score = -1.57, R) >droYak2.chr3R 6982789 91 + 28832112 ---------UUUGUGUUCCUCCUCCGCCGAGCAAAUCCCACAGCUUGAUGAAGCACUUCCUGCGGCUCAAGUGUCGCCCA----UCGUGUGCGAAUUUCGCUCG ---------..................(((((((((.(((((((((....)))).......(((((......)))))...----...)))).).)))).))))) ( -23.10, z-score = -0.99, R) >droEre2.scaffold_4820 7088396 100 + 10470090 UUUUUGUUCCUCCUCCUCCGCUUUCGCCGAGCAUAUCCCACAGCUUGAUGAAGCACUUCCUGCGGCUCAAGUGUCGCCCA----UCGUGUGCGAAUUUCGCUCG ...................((.((((((((((..........))))).....((((.....(((((......)))))...----..)))))))))....))... ( -22.70, z-score = -0.80, R) >droAna3.scaffold_13340 17451220 86 - 23697760 ------------CCCAAACACUUGCCCCGCCUCUGGAAUA-AGCUUGAUGAAGCACUUCCUGCGGCUCAAGUGUCGCCCAUGGGUUAUGUGCGAAAAUG----- ------------......((((((..((((....((((..-.((((....))))..)))).))))..))))))((((.((((...)))).)))).....----- ( -27.30, z-score = -2.45, R) >droGri2.scaffold_15074 3825115 81 - 7742996 ----------------UAUAUAUAUA-AGAGUGAAUAACGCGGCAUGAUUAAUU-UUGCGUAGGCGAUGAACGUCGCGU-----UUUUCUGCAAUUGAAACACA ----------------..........-...(((....((((((.((.....)).-))))))..(((((....)))))..-----.((((.......))))))). ( -15.80, z-score = 0.31, R) >consensus UUU_UGUUCCUCC_CCAACGCCUUCACCGAGCAAAUCCCACAGCUUGAUGAAGCACUUCCUGCGGCUCAAGUGUCGCCCA____UCGUGUGCGAAUUUCACUCG ............................(((...........((((((.(..(((.....)))..))))))).((((.((.......)).))))......))). (-10.55 = -11.03 + 0.47)

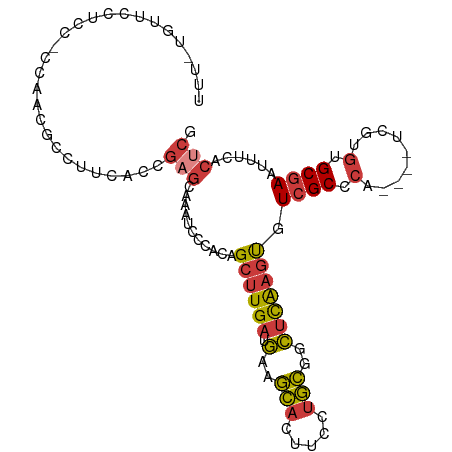

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:52:37 2011