| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,651,860 – 24,651,952 |
| Length | 92 |
| Max. P | 0.951799 |

| Location | 24,651,860 – 24,651,952 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 61.47 |
| Shannon entropy | 0.75270 |
| G+C content | 0.43201 |
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -10.56 |
| Energy contribution | -10.86 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.951799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
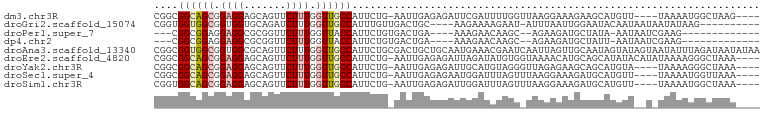
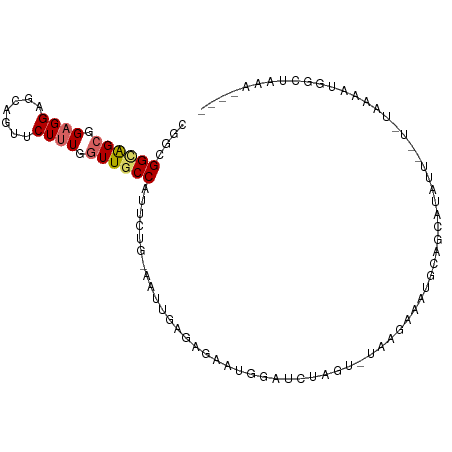
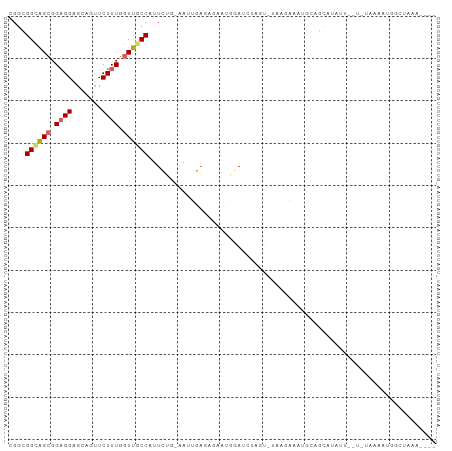
>dm3.chr3R 24651860 92 + 27905053 CGGCGGCAGCGGAGGAGCAGUUCUUUGGUUGCCAUUCUG-AAUUGAGAGAUUCGAUUUUGGUUAAGGAAAGAAGCAUGUU----UAAAAUGGCUAAG---- .(((((((((.((((((...)))))).)))))).((((.-(((..(((.......)))..))).))))............----.......)))...---- ( -24.90, z-score = -1.32, R) >droGri2.scaffold_15074 3818802 86 + 7742996 CGGUGGUGGCGGUGGUGCAGAUCUUUGGUUGCCAUUUGUUGACUGC----AAGAAAAGAAU-AUUUAAUUGGAAUACAAUAAUAAUAUAAG---------- ..((((..((.(.((.......)).).))..))))((((.....))----)).......((-(((..((((.....))))...)))))...---------- ( -14.20, z-score = 0.45, R) >droPer1.super_7 974102 78 + 4445127 ---CGGCGGAGGAGGCGCGGUUCUUUGGUUACCAUUCUGUGACUGA----AAAGAACAAGC--AGAAGAUGCUAUA-AAUAAUCGAAG------------- ---.((((......((...(((((((((((((......))))))..----)))))))..))--......))))...-...........------------- ( -19.30, z-score = -1.24, R) >dp4.chr2 27426929 78 - 30794189 ---CGGCGGAGGAGGCGCGGUUCUUUGGUUACCAUUCUGUGACUGA----AAAGAACAAGC--AGAAGAUGCUAUU-AAUAAUCGAAG------------- ---.((((......((...(((((((((((((......))))))..----)))))))..))--......))))...-...........------------- ( -19.30, z-score = -1.11, R) >droAna3.scaffold_13340 17447474 101 + 23697760 CGGCGGUGGCGGUGGCGCAGUUCUUUGGUUGCCAUUCUGCGACUGCUGCAAUGAAACGAAUCAAUUAGUUGCAAUAGUAUAGUAAUAUUUAGAUAAUAUAA .((((((.((((((((((((....)))..)))))..)))).))))))(((((...............)))))............(((((.....))))).. ( -24.86, z-score = -0.00, R) >droEre2.scaffold_4820 7084167 96 - 10470090 CGGCGGCAGCGGAGGAGCAGUUCUUUGGUUGCCAUUCUG-AAUUGAGAGAUUAGAUAUGUGGUAAAACAUGCAGCAUAUACAUAUAAAAGGGCUAAA---- .(((((((((.((((((...)))))).))))))..((((-(.((....)))))))(((((.(((.....))).))))).............)))...---- ( -26.30, z-score = -2.04, R) >droYak2.chr3R 6978531 92 - 28832112 CGGCGGCAGCGGAGGAGCAGUUCUUUGGUUGCCAUUCUG-AAUUGAGAGAUUGCAUGUAGGGUUAGAGAAGCAGCAUGUA----UAAAAGGGCUAAA---- .(((((((((.((((((...)))))).))))))..(((.-((((....))))((((((...(((.....))).)))))).----....))))))...---- ( -26.80, z-score = -1.74, R) >droSec1.super_4 3515339 92 + 6179234 CGGCGGCAGCGGAGGAGCAGUUCUUUGGUUGCCAUUCUG-AAUUGAGAGAAUGGAUUUAGUUUAAGGAAAGAUGCAUGUU----UAAAAUGGUUAAA---- ..((((((((.((((((...)))))).))))))...(((-((((.(.....).))))))).............)).....----.............---- ( -22.20, z-score = -1.33, R) >droSim1.chr3R 24333105 92 + 27517382 CGGUGGCAGCGGAGGAGCAGUUCUUUGGUUGCCAUUCUG-AAUUGAGAGAUUGGAUUUAGUUUAAGGAAAGAUGCAUGUU----UAAAAUGGCUAAA---- ..((((((((.((((((...)))))).))))))))(((.-((((....)))))))(((((((.....(((.(....).))----).....)))))))---- ( -23.00, z-score = -1.35, R) >consensus CGGCGGCAGCGGAGGAGCAGUUCUUUGGUUGCCAUUCUG_AAUUGAGAGAAUGGAUCUAGU_UAAGAAAUGCAGCAUAUU__U_UAAAAUGGCUAAA____ ....((((((.((((.......)))).)))))).................................................................... (-10.56 = -10.86 + 0.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:52:36 2011