
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,604,560 – 24,604,670 |
| Length | 110 |
| Max. P | 0.843272 |

| Location | 24,604,560 – 24,604,669 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 86.75 |
| Shannon entropy | 0.29250 |
| G+C content | 0.42866 |
| Mean single sequence MFE | -26.89 |
| Consensus MFE | -18.35 |
| Energy contribution | -18.18 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.707984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24604560 109 - 27905053 ----GCACCACCCAGUGCACCAGCACUCGUGGCUGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUAUUUUGCACUCG ----(((((((..(((((....))))).)))).((((((.((((.(((((.(((((......))))))))))..)))).)))))).)))(((((((....)))))))...... ( -30.30, z-score = -2.42, R) >droSim1.chr3R 24290459 109 - 27517382 ----GCACCACCCAGUGCACCAGCACUCGUGGCUGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUAUUUUGCACUCG ----(((((((..(((((....))))).)))).((((((.((((.(((((.(((((......))))))))))..)))).)))))).)))(((((((....)))))))...... ( -30.30, z-score = -2.42, R) >droSec1.super_4 3473401 109 - 6179234 ----GCACCACCCAGUGCACCAGCACUCGUGGCUGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUAUUUUGCACUCG ----(((((((..(((((....))))).)))).((((((.((((.(((((.(((((......))))))))))..)))).)))))).)))(((((((....)))))))...... ( -30.30, z-score = -2.42, R) >droYak2.chr3R 6934970 113 + 28832112 CUCGGCACCACCCAGUGCACCAGCACUCGUGGCUGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUAUUUUGCACUCG ...((((((((..(((((....))))).)))).((((((.((((.(((((.(((((......))))))))))..)))).)))))).))))...((((.......))))..... ( -32.80, z-score = -2.69, R) >droEre2.scaffold_4820 7040736 111 + 10470090 --CCGCACCACCCAGUGCACCAGCACUCGUGGCUGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUAUUUUGCACUCG --..(((((((..(((((....))))).)))).((((((.((((.(((((.(((((......))))))))))..)))).)))))).)))(((((((....)))))))...... ( -30.60, z-score = -2.44, R) >dp4.chr2 27380608 104 + 30794189 ----UAACCGCCC--UGCACCA---CUCGUGGCUGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUAUUUUGCACUCG ----.........--((((...---....((((((((((.((((.(((((.(((((......))))))))))..)))).))))))..))))(((((....))))))))).... ( -22.80, z-score = -1.24, R) >droPer1.super_7 926750 104 - 4445127 ----UAACCGCCC--UGCACCA---CUCGUGGCUGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUAUUUUGCACUCG ----.........--((((...---....((((((((((.((((.(((((.(((((......))))))))))..)))).))))))..))))(((((....))))))))).... ( -22.80, z-score = -1.24, R) >droWil1.scaffold_181089 9310749 102 - 12369635 ----GGGCCUCACAUUGCGC-------UGUGGCUGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUAUUUUGCACUCG ----.((((.((........-------)).))))(((((.((((.(((((.(((((......))))))))))..)))).)))))..(((.((((((....))))))))).... ( -24.50, z-score = -0.68, R) >droVir3.scaffold_13047 8935882 96 + 19223366 ---------------UGCCCCUG--GUGGUAGCUGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUAUUUUGCACUCG ---------------(((.....--..(((((.((((((.((((.(((((.(((((......))))))))))..)))).)))))))))))((((((....))))))))).... ( -23.40, z-score = -1.54, R) >droMoj3.scaffold_6540 5715786 98 + 34148556 ---------------UGCCCCUUGGGUGGCAGUCGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUAUUUUGCACUCG ---------------........((((((((((.(((((.((((.(((((.(((((......))))))))))..)))).))))))))))(((((((....)))))))))))). ( -28.30, z-score = -2.55, R) >droGri2.scaffold_14624 4198244 105 + 4233967 --------UGCCCCGUCUAGUCGUUGUGUUAGUGGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCACAUUGCCGAAGUAACUUUAUUUUGCUCUCC --------.((.......(((..((.((.(((((.((((.((((.(((((.(((((......))))))))))..)))).))))))))).)).))..))).......))..... ( -19.64, z-score = -0.62, R) >consensus ____GCACCACCCAGUGCACCAGCACUCGUGGCUGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUAUUUUGCACUCG ............................((((.((((((.((((.(((((.(((((......))))))))))..)))).))))))))))(((((((....)))))))...... (-18.35 = -18.18 + -0.17)

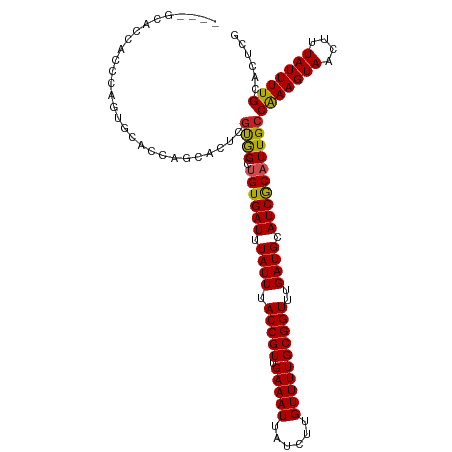

| Location | 24,604,571 – 24,604,670 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.19 |
| Shannon entropy | 0.35596 |
| G+C content | 0.43342 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -15.70 |
| Energy contribution | -15.61 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.843272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24604571 99 - 27905053 --------------GGCACCACCCAGUGCACCAGCACUCGUGGCUGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUA --------------((((((((..(((((....))))).)))).((((((.((((.(((((.(((((......))))))))))..)))).)))))).))))............ ( -31.90, z-score = -3.03, R) >droSim1.chr3R 24290470 99 - 27517382 --------------GGCACCACCCAGUGCACCAGCACUCGUGGCUGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUA --------------((((((((..(((((....))))).)))).((((((.((((.(((((.(((((......))))))))))..)))).)))))).))))............ ( -31.90, z-score = -3.03, R) >droSec1.super_4 3473412 99 - 6179234 --------------GGCACCACCCAGUGCACCAGCACUCGUGGCUGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUA --------------((((((((..(((((....))))).)))).((((((.((((.(((((.(((((......))))))))))..)))).)))))).))))............ ( -31.90, z-score = -3.03, R) >droYak2.chr3R 6934981 113 + 28832112 GGCCCCACCCACUCGGCACCACCCAGUGCACCAGCACUCGUGGCUGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUA ((......))....((((((((..(((((....))))).)))).((((((.((((.(((((.(((((......))))))))))..)))).)))))).))))............ ( -32.50, z-score = -2.12, R) >droEre2.scaffold_4820 7040747 111 + 10470090 --GGCACCACCCACCGCACCACCCAGUGCACCAGCACUCGUGGCUGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUA --((((............((((..(((((....))))).)))).((((((.((((.(((((.(((((......))))))))))..)))).)))))).))))............ ( -31.90, z-score = -2.31, R) >dp4.chr2 27380619 93 + 30794189 ---------------UAACCGCCC--UGCACCA---CUCGUGGCUGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUA ---------------.........--......(---((..((((((((((.((((.(((((.(((((......))))))))))..)))).))))))..)))).)))....... ( -21.90, z-score = -1.55, R) >droPer1.super_7 926761 93 - 4445127 ---------------UAACCGCCC--UGCACCA---CUCGUGGCUGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUA ---------------.........--......(---((..((((((((((.((((.(((((.(((((......))))))))))..)))).))))))..)))).)))....... ( -21.90, z-score = -1.55, R) >droWil1.scaffold_181089 9310760 92 - 12369635 --------------GGGGCCUCACAUUGCGC-------UGUGGCUGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUA --------------...((........))((-------(.((((((((((.((((.(((((.(((((......))))))))))..)))).))))))..)))).)))....... ( -23.90, z-score = -0.82, R) >droVir3.scaffold_13047 8935893 85 + 19223366 -------------------UGCCC---CUG------GUGGUAGCUGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUA -------------------.....---((.------.((((((.((((((.((((.(((((.(((((......))))))))))..)))).)))))))))))).))........ ( -22.90, z-score = -2.25, R) >droMoj3.scaffold_6540 5715797 87 + 34148556 -------------------UGCCCC--UUGG-----GUGGCAGUCGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUA -------------------.((((.--..))-----))((((((.(((((.((((.(((((.(((((......))))))))))..)))).)))))))))))............ ( -27.20, z-score = -2.93, R) >droGri2.scaffold_14624 4198255 94 + 4233967 -------------------UGCCCCGUCUAGUCGUUGUGUUAGUGGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCACAUUGCCGAAGUAACUUUA -------------------..............((((..(..((.(((((.((((.(((((.(((((......))))))))))..)))).)))))...))..)..)))).... ( -18.80, z-score = -0.57, R) >consensus _______________GCACCACCCAGUGCACCAG__CUCGUGGCUGUGAUUUAUUUACCGUUGAAAUUAUCUUGUUUUGCGGUUUGAUGCAUCGCAUUGCCAAAGUAACUUUA .......................................((((.((((((.((((.(((((.(((((......))))))))))..)))).))))))))))............. (-15.70 = -15.61 + -0.09)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:52:30 2011