| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,555,392 – 24,555,503 |
| Length | 111 |
| Max. P | 0.901465 |

| Location | 24,555,392 – 24,555,503 |
|---|---|
| Length | 111 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 62.86 |
| Shannon entropy | 0.77881 |
| G+C content | 0.41358 |
| Mean single sequence MFE | -21.53 |
| Consensus MFE | -7.80 |
| Energy contribution | -8.48 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.79 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.901465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
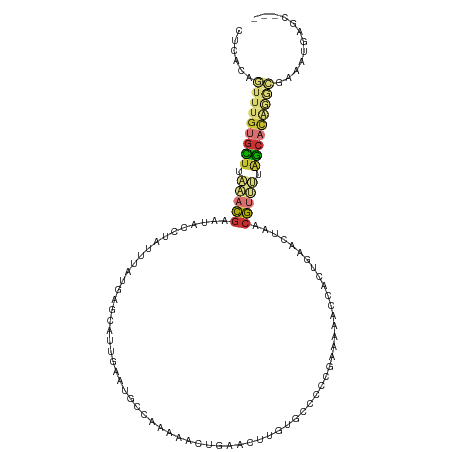
>dm3.chr3R 24555392 111 - 27905053 UUCACAGUUUGUGCUUAAACGAAUACCUAUUUAUGAGCAUUGAAUGCCAAAAAUUGAACUUGUGCCCCCGAAAAACCACUGAACUAACGUUUUAGCACAGGCGAAAUGAGC--- ((((..(((((((((.(((((.....(.((((.((.((((...)))))).)))).).....(((............)))........))))).)))))))))....)))).--- ( -22.60, z-score = -2.04, R) >droSim1.chr3R 24254132 111 - 27517382 CUCACAGUUUGUGCCUAAACGAAUACCUAUUUAUGAGCAUUGAAUGCCAAAAACUGAACUUGUGCCCGCGAAAAACCACUGAACUAACGUUUUAGCACAGGCGAAAUGAGC--- ((((..((((((((..(((((............((.((((...))))))..........((((....))))................)))))..))))))))....)))).--- ( -23.90, z-score = -1.99, R) >droSec1.super_4 3436976 111 - 6179234 CUCACAGUUUGUGCUUAAACGAAUACCUAUUUAUGAGCAUUGAAUGCCAAAAACUGAACUUGUGCCCCCGAAAAACCACUGAACUAACGUUUUAGCACAGGCGAAGUGAGC--- (((((.(((((((((.(((((............((.((((...))))))............(((............)))........))))).)))))))))...))))).--- ( -26.80, z-score = -2.88, R) >droYak2.chr3R 6898201 111 + 28832112 CGCACAGUUUGUGCUUAAACGAAUACCUAUUUAUGAGCAUUGAAUGCCAAAAACUGAACUUGUGCACCCGAAAAACCAGUGAACUAACGUUUUAGCACAGGCGAAAUGAGC--- ......(((((((((.(((((..................(((..((((((.........))).)))..)))......((....))..))))).))))))))).........--- ( -23.20, z-score = -1.13, R) >droEre2.scaffold_4820 7002363 110 + 10470090 CUCACAGUUUGUGCUUAAACG-AUACCUAUUUAUGAGCAUGGAAUGCCAAAAACUGAACUUGUGCCCCCGAAAAACCACUGAACUAACGUUUUAGCACAGGCGAAAUGAGU--- (((((((((((((((((...(-((....)))..)))))))((....))..))))))..(((((((....(......)...((((....))))..))))))).....)))).--- ( -24.50, z-score = -2.14, R) >droAna3.scaffold_13340 17361836 102 - 23697760 CUUGCAUUUUGCGUUU---UGAAUAGAACCCAAUUUAUGUCA-----CAAAACGCGAACUUGUGCGCCC-AAACGCGAGGGAGCCAACAUUUUAGCAUGAAAGCCAUGAGC--- ..(((..(((((((((---((.(((((......)))))....-----))))))))))).(((.((.(((-........))).))))).......)))..............--- ( -24.60, z-score = -0.91, R) >dp4.chr2 27340916 98 + 30794189 -------AAUGUGCUC---UGCACUCCACUCUAUU--UUCUGCA---CAAAAACUAAACUUUUGACCCCCGAAAUUUGGCCGGCUAACAUUUUAGCAUUAA-GAAACGAGCCAU -------.(((.((((---((((............--...))))---......(((((.(((((.....))))))))))...(((((....))))).....-.....))))))) ( -15.36, z-score = -0.72, R) >droWil1.scaffold_181089 9269569 91 - 12369635 -------CUAGCAUUCUCACUAAAAACAAAACA-GAACAGCGAGAGGCGACAAAAAAAUCUGCGUAAAUGACAUUUAUUCUAGUCAGUGUGAAGACAAC--------------- -------...(..(((.((((............-.....((((((.............))).)))....(((..........))))))).)))..)...--------------- ( -11.32, z-score = 0.71, R) >consensus CUCACAGUUUGUGCUUAAACGAAUACCUAUUUAUGAGCAUUGAAUGCCAAAAACUGAACUUGUGCCCCCGAAAAACCACUGAACUAACGUUUUAGCACAGGCGAAAUGAGC___ ......(((((((((.(((((..................................................................))))).)))))))))............ ( -7.80 = -8.48 + 0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:52:23 2011