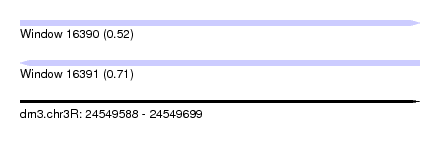
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,549,588 – 24,549,699 |
| Length | 111 |
| Max. P | 0.713718 |

| Location | 24,549,588 – 24,549,699 |
|---|---|
| Length | 111 |
| Sequences | 13 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.68 |
| Shannon entropy | 0.37511 |
| G+C content | 0.49827 |
| Mean single sequence MFE | -35.97 |
| Consensus MFE | -22.56 |
| Energy contribution | -22.35 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.63 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.516226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24549588 111 + 27905053 UUCAUGACUUGCCUGCUGCUGAUCAACAAGCUGUACGACGAGCACAUGGUCGAUCAGCAGGAGCAGUUCCAGAUCAAGCAGGUCUGCCUCCAGGCCAUCCUCGAACUUUUA (((.......(((((((..(((((.....((((..((((.(.....).))))..)))).((((...)))).))))))))))))..(((....))).......)))...... ( -34.60, z-score = -0.89, R) >droSim1.chr3R 24248352 111 + 27517382 UUCAUGACCUGCCUGCUGCUGAUCAACAAGCUGUACGACGAGCACAUGGUCGAUCAGCAGGAGCAGUUCCAGAUCAAGCAGGUCUGCCUCCAGGCCAUCCUCGAACUUUUA (((..(((((((((.(((((((((.((.....))..(((.(.....).)))))))))))).))..(........)..))))))).(((....))).......)))...... ( -36.10, z-score = -1.29, R) >droSec1.super_4 3431186 111 + 6179234 UUCAUGACCUGCCUGCUGCUGAUCAACAAGCUGUACGACGAACACAUGGUCGAUCAGCAGGAGCAGUUCCAGAUCAAGCAGGUCUGCCUCCAGGCCAUCCUCGAACUUUUA (((..(((((((((.(((((((((.((.....))..(((.(.....).)))))))))))).))..(........)..))))))).(((....))).......)))...... ( -35.30, z-score = -1.56, R) >droYak2.chr3R 6892330 111 - 28832112 UUCAUGACCUGCCUGCUGCUGAUCAACAAGCUGUACGACGAGCACAUGGUCGAUCAGCAGGAACAGUUCCAGAUCAAGCAGGUCUGCCUCCAGGCCAUCCUCGAACUUUUA (((..(((((((.((.((((((((.((.....))..(((.(.....).)))))))))))(((.....)))....)).))))))).(((....))).......)))...... ( -35.60, z-score = -1.65, R) >droEre2.scaffold_4820 6996504 111 - 10470090 UUCAUGACCUGCCUGCUGCUGAUCAACAAGCUGUACGACGAGCACAUGGUCGAUCAGCAGGAGCAGUUCCAGAUCAAGCUGGUCUGCCUCCAGGCCAUCCUCGAACUUUUA ..........((((((((((((((.((.....))..(((.(.....).))))))))))))((((((..((((......)))).))).))))))))................ ( -37.70, z-score = -1.66, R) >droAna3.scaffold_13340 17355336 111 + 23697760 UUCAUGACCUGCCUCCUGCUGAUCAAUAAGCUGUACGAUGAGCACAUGGUCGAUCAGCAGGAACAGUUCCAGAUCAAGCAGGUCUGCCUCCAGGCCAUCCUCGAACUUUUA (((..(((((((.(((((((((((.((..(((........))).....)).)))))))))))...(........)..))))))).(((....))).......)))...... ( -36.60, z-score = -2.29, R) >dp4.chr2 27334470 111 - 30794189 UUUAUGACUUUCCUGCUGCUGAUCAACAAGGUGUACGACGAGCACAUGGUCGAUCAGCAGGAGCAGUUCCAGAUCAAGCAGGUCUGCCUGCAGGCCAUCAUGGAACUUUUA ............((.(((((((((.((...((((.......))))...)).))))))))).)).(((((((((((..(((((....)))))..)..))).))))))).... ( -42.80, z-score = -3.24, R) >droPer1.super_7 880491 111 + 4445127 UUUAUGACUUUCCUGCUGCUGAUCAACAAGGUGUACGACGAGCACAUGGUCGAUCAGCAGGAGCAGUUCCAGAUCAAGCAGGUCUGCCUGCAGGCCAUCAUGGAACUUUUA ............((.(((((((((.((...((((.......))))...)).))))))))).)).(((((((((((..(((((....)))))..)..))).))))))).... ( -42.80, z-score = -3.24, R) >droWil1.scaffold_181089 9263685 111 + 12369635 UUCAUGACAUUUCUGUUGCUUAUUAAUAAAGUGUAUGAUGAGCAUAUGGUCGAUCAUCAGGAGCAAUAUCAGAUCAAGCAAGUCUGUCUGCUGGCCAUUAUGGAACUUUUA (((((((.........(((((((((..........)))))))))..(((((((((.....((......)).)))).((((........))))))))))))))))....... ( -24.70, z-score = 0.22, R) >droVir3.scaffold_13047 8875199 111 - 19223366 UUUAUGACCUUUCUGCUGCUGAUCAACAAGGAGUACGAUGAGCAUAUGGUCGAUCAGCAGGAACAGUUCCAGAUUAAACAGGUCUGCCUGCUGGCCAUCAUGGAGCUGCUA ...............(((((((((.....(.....)(((.(.....).))))))))))))...(((((((((((....((((....))))......))).))))))))... ( -34.70, z-score = -0.90, R) >droMoj3.scaffold_6540 5664795 111 - 34148556 UUCAUGACAUUCCUGCUGCUGAUCAACAAGGUGUACGAUGAGCACAUGGUCGAUCAACAGGAACAGUUCCAGAUCAAGCAGGUCUGCCUGCAGGCCAUCAUGGAGCUGCUG .........((((((....(((((.((...((((.......))))...)).))))).))))))((((((((((((..(((((....)))))..)..))).))))))))... ( -39.50, z-score = -1.53, R) >droGri2.scaffold_14624 4162894 111 - 4233967 UUUAUGACAUUUCUGCUGUUAAUCAAUAAGCUGUACGAUGAGCACAUGGUCGAUCAGCAGGAGCAAUUCCAGAUCAAGCAGCUCUGCUUGCAGGCCAUCAUGGAGCUGCUA ..........(((((((((..((((....(((........)))...))))..).))))))))(((.((((((((((((((....))))))......))).))))).))).. ( -32.90, z-score = -0.29, R) >anoGam1.chr2R 43854638 111 + 62725911 UUUAUGUCACUGAUGGUGCUGCUGAACCGCUCGGAGGAUCCCAUUUUCGUGAGCCUGGUCGAGCAGCGGGAAAUCAAAAACAUGUGCCUGCAGGCUGUAAUGUCGCUGCUG ..(((((...((((..(((((((..((((((((((((((...)))))).)))))..)))..)))))))....))))...))))).((..((.(((......))))).)).. ( -34.30, z-score = -0.06, R) >consensus UUCAUGACCUUCCUGCUGCUGAUCAACAAGCUGUACGACGAGCACAUGGUCGAUCAGCAGGAGCAGUUCCAGAUCAAGCAGGUCUGCCUGCAGGCCAUCAUGGAACUUUUA ...............(((((((((.((....(((.......)))....)).)))))))))....(((((.(((((.....)))))(((....))).......))))).... (-22.56 = -22.35 + -0.21)

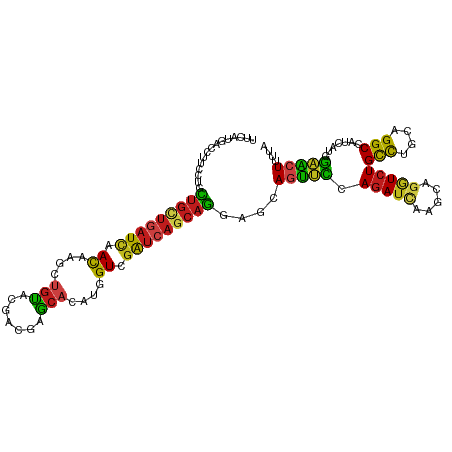
| Location | 24,549,588 – 24,549,699 |
|---|---|
| Length | 111 |
| Sequences | 13 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.68 |
| Shannon entropy | 0.37511 |
| G+C content | 0.49827 |
| Mean single sequence MFE | -40.41 |
| Consensus MFE | -17.76 |
| Energy contribution | -18.74 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.713718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24549588 111 - 27905053 UAAAAGUUCGAGGAUGGCCUGGAGGCAGACCUGCUUGAUCUGGAACUGCUCCUGCUGAUCGACCAUGUGCUCGUCGUACAGCUUGUUGAUCAGCAGCAGGCAAGUCAUGAA ......((((.((...(((((..(((((.((..........))..))))).((((((((((((..(((((.....)))))....)))))))))))))))))...)).)))) ( -42.40, z-score = -1.99, R) >droSim1.chr3R 24248352 111 - 27517382 UAAAAGUUCGAGGAUGGCCUGGAGGCAGACCUGCUUGAUCUGGAACUGCUCCUGCUGAUCGACCAUGUGCUCGUCGUACAGCUUGUUGAUCAGCAGCAGGCAGGUCAUGAA .....((((.(((....))).)).)).((((((((((....(((.....)))(((((((((((..(((((.....)))))....))))))))))).))))))))))..... ( -48.30, z-score = -3.30, R) >droSec1.super_4 3431186 111 - 6179234 UAAAAGUUCGAGGAUGGCCUGGAGGCAGACCUGCUUGAUCUGGAACUGCUCCUGCUGAUCGACCAUGUGUUCGUCGUACAGCUUGUUGAUCAGCAGCAGGCAGGUCAUGAA .....((((.(((....))).)).)).((((((((((....(((.....)))(((((((((((..((((.......))))....))))))))))).))))))))))..... ( -44.50, z-score = -2.49, R) >droYak2.chr3R 6892330 111 + 28832112 UAAAAGUUCGAGGAUGGCCUGGAGGCAGACCUGCUUGAUCUGGAACUGUUCCUGCUGAUCGACCAUGUGCUCGUCGUACAGCUUGUUGAUCAGCAGCAGGCAGGUCAUGAA .....((((.(((....))).)).)).((((((((((....(((.....)))(((((((((((..(((((.....)))))....))))))))))).))))))))))..... ( -48.10, z-score = -3.51, R) >droEre2.scaffold_4820 6996504 111 + 10470090 UAAAAGUUCGAGGAUGGCCUGGAGGCAGACCAGCUUGAUCUGGAACUGCUCCUGCUGAUCGACCAUGUGCUCGUCGUACAGCUUGUUGAUCAGCAGCAGGCAGGUCAUGAA .............((((((((..(((((.((((......))))..))))).((((((((((((..(((((.....)))))....))))))))))))....))))))))... ( -46.30, z-score = -2.70, R) >droAna3.scaffold_13340 17355336 111 - 23697760 UAAAAGUUCGAGGAUGGCCUGGAGGCAGACCUGCUUGAUCUGGAACUGUUCCUGCUGAUCGACCAUGUGCUCAUCGUACAGCUUAUUGAUCAGCAGGAGGCAGGUCAUGAA .....((((.(((....))).)).)).((((((((..............(((((((((((((...(((((.....))))).....)))))))))))))))))))))..... ( -45.03, z-score = -3.11, R) >dp4.chr2 27334470 111 + 30794189 UAAAAGUUCCAUGAUGGCCUGCAGGCAGACCUGCUUGAUCUGGAACUGCUCCUGCUGAUCGACCAUGUGCUCGUCGUACACCUUGUUGAUCAGCAGCAGGAAAGUCAUAAA ....(((((((.(((.(...(((((....))))).).)))))))))).((.((((((((((((..(((((.....)))))....)))))))))))).))............ ( -44.40, z-score = -4.23, R) >droPer1.super_7 880491 111 - 4445127 UAAAAGUUCCAUGAUGGCCUGCAGGCAGACCUGCUUGAUCUGGAACUGCUCCUGCUGAUCGACCAUGUGCUCGUCGUACACCUUGUUGAUCAGCAGCAGGAAAGUCAUAAA ....(((((((.(((.(...(((((....))))).).)))))))))).((.((((((((((((..(((((.....)))))....)))))))))))).))............ ( -44.40, z-score = -4.23, R) >droWil1.scaffold_181089 9263685 111 - 12369635 UAAAAGUUCCAUAAUGGCCAGCAGACAGACUUGCUUGAUCUGAUAUUGCUCCUGAUGAUCGACCAUAUGCUCAUCAUACACUUUAUUAAUAAGCAACAGAAAUGUCAUGAA .....(((((.....))..))).((((...(((((((...(((((..(....((((((.((......)).))))))....)..))))).)))))))......))))..... ( -18.30, z-score = 0.44, R) >droVir3.scaffold_13047 8875199 111 + 19223366 UAGCAGCUCCAUGAUGGCCAGCAGGCAGACCUGUUUAAUCUGGAACUGUUCCUGCUGAUCGACCAUAUGCUCAUCGUACUCCUUGUUGAUCAGCAGCAGAAAGGUCAUAAA .(((((.((((.(((....((((((....))))))..))))))).))))).((((((((((((....(((.....)))......))))))))))))............... ( -36.80, z-score = -2.16, R) >droMoj3.scaffold_6540 5664795 111 + 34148556 CAGCAGCUCCAUGAUGGCCUGCAGGCAGACCUGCUUGAUCUGGAACUGUUCCUGUUGAUCGACCAUGUGCUCAUCGUACACCUUGUUGAUCAGCAGCAGGAAUGUCAUGAA ..((((..((.....)).)))).((((..(((((.......(((.....)))(((((((((((..(((((.....)))))....))))))))))))))))..))))..... ( -41.60, z-score = -2.06, R) >droGri2.scaffold_14624 4162894 111 + 4233967 UAGCAGCUCCAUGAUGGCCUGCAAGCAGAGCUGCUUGAUCUGGAAUUGCUCCUGCUGAUCGACCAUGUGCUCAUCGUACAGCUUAUUGAUUAACAGCAGAAAUGUCAUAAA .(((((.((((.(((......((((((....))))))))))))).))))).(((((((((((...(((((.....))))).....)))))...))))))............ ( -32.90, z-score = -0.81, R) >anoGam1.chr2R 43854638 111 - 62725911 CAGCAGCGACAUUACAGCCUGCAGGCACAUGUUUUUGAUUUCCCGCUGCUCGACCAGGCUCACGAAAAUGGGAUCCUCCGAGCGGUUCAGCAGCACCAUCAGUGACAUAAA ..(((((.........).))))......(((((.(((((.....((((((.((((..((((.(......)((.....)))))))))).))))))...))))).)))))... ( -32.30, z-score = -0.95, R) >consensus UAAAAGUUCCAUGAUGGCCUGCAGGCAGACCUGCUUGAUCUGGAACUGCUCCUGCUGAUCGACCAUGUGCUCGUCGUACAGCUUGUUGAUCAGCAGCAGGAAGGUCAUGAA .............(((((....(((((....))))).........(((...(((((((((((.......................))))))))))))))....)))))... (-17.76 = -18.74 + 0.98)
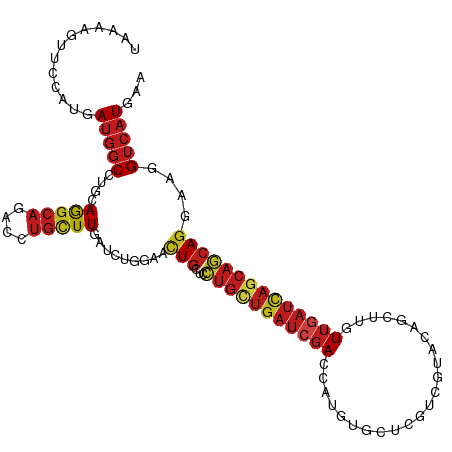
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:52:19 2011