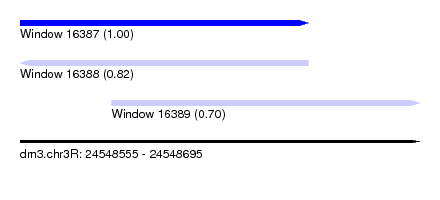
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,548,555 – 24,548,695 |
| Length | 140 |
| Max. P | 0.996939 |

| Location | 24,548,555 – 24,548,656 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.41 |
| Shannon entropy | 0.36729 |
| G+C content | 0.57485 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -17.15 |
| Energy contribution | -18.54 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.68 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.996939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
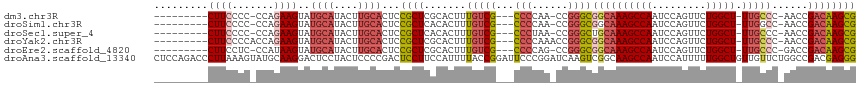
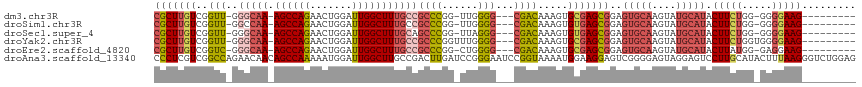
>dm3.chr3R 24548555 101 + 27905053 ---------CUUCCCC-CCAGAAGUAUGCAUACUUGCACUCCGCUCGCACUUUGUCG---CCCCAA-CCGGGCGGCAAAGCCAAUCCAGUUCUGGCU-UUGCCC-AACCGACAAGCG ---------((((...-...))))..((((....)))).......(((...((((((---(((...-..))))((((((((((.........)))))-))))).-....)))))))) ( -36.20, z-score = -4.46, R) >droSim1.chr3R 24247316 101 + 27517382 ---------CUUCCCC-CCAGAAGUAUGCAUACUUGCACUCCGCUCACACUUUGUCG---CCCCAA-CCGGGCGGCAAAGCCAAUCCAGUUCUGGCU-UUGGCC-AACCGACAAGCG ---------((((...-...))))..((((....))))...((((.....(((((((---(((...-..))))))))))(((((.(((....)))..-))))).-........)))) ( -34.10, z-score = -3.46, R) >droSec1.super_4 3430166 101 + 6179234 ---------CUUCCCC-CCAGAAGUAUGCAUACUUGCACUCCGCUCACACUUUGUCG---CCCUAA-CCGGGCUGCAAAGCCAAUCCAGUUCUGGCU-UUGCCC-AACCGACAAGCG ---------((((...-...))))..((((....))))...((((.......(((((---(((...-..)))).(((((((((.........)))))-))))..-....)))))))) ( -32.11, z-score = -4.24, R) >droYak2.chr3R 6891230 103 - 28832112 ---------CUUCCCCACCAGAAGUAUGCAUACUUGCACUCCGCUCGCACUUUGUCG---CCCCAAACCGGGCGGCAAAGCCAAUCCAGUUCUGGCU-UUGCCC-AACCGACAAGCG ---------((((.......))))..((((....)))).......(((...((((((---(((......))))((((((((((.........)))))-))))).-....)))))))) ( -35.90, z-score = -4.33, R) >droEre2.scaffold_4820 6995447 101 - 10470090 ---------CUUCCUC-CCAUAAGUAUGCAUACUUGCACUCCGCUCGCACUUUGUCG---CCCCAG-CCGGGCGGCAAAGCCAAUCCAGUUCUGGCU-UUGCCC-GACCGACAAGCG ---------.......-.........((((....)))).......(((...((((((---(((...-..))))((((((((((.........)))))-))))).-....)))))))) ( -34.60, z-score = -3.34, R) >droAna3.scaffold_13340 17354262 117 + 23697760 CUCCAGACCCUUAAAGUAUGCAAGGACUCCUACUCCCCGACUCCUUCCAUUUUACCGGAUUCCCGGAUCAAGUCGGCAAGCCAAUCCAUUUUUGGCUGUUGUUCUGGCCGACGAGGG .......(((((.....(((.(((((.((.........)).))))).)))....((((....)))).....((((((.((((((.......)))))).........))))))))))) ( -36.90, z-score = -2.26, R) >consensus _________CUUCCCC_CCAGAAGUAUGCAUACUUGCACUCCGCUCGCACUUUGUCG___CCCCAA_CCGGGCGGCAAAGCCAAUCCAGUUCUGGCU_UUGCCC_AACCGACAAGCG ..........................((((....))))...((((.......(((((....(((.....))).((((((((((.........))))).))))).....))))))))) (-17.15 = -18.54 + 1.39)
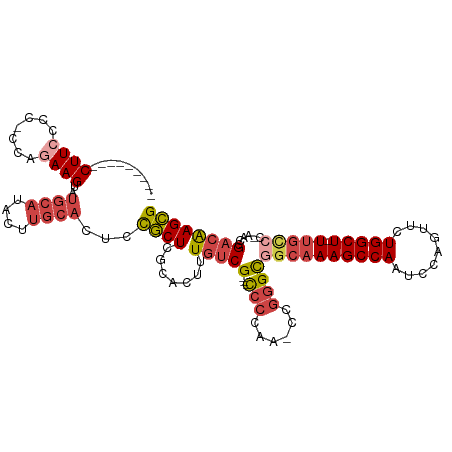
| Location | 24,548,555 – 24,548,656 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.41 |
| Shannon entropy | 0.36729 |
| G+C content | 0.57485 |
| Mean single sequence MFE | -41.52 |
| Consensus MFE | -22.47 |
| Energy contribution | -23.98 |
| Covariance contribution | 1.51 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.824236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24548555 101 - 27905053 CGCUUGUCGGUU-GGGCAA-AGCCAGAACUGGAUUGGCUUUGCCGCCCGG-UUGGGG---CGACAAAGUGCGAGCGGAGUGCAAGUAUGCAUACUUCUGG-GGGGAAG--------- (((((((...((-((((((-((((((.......)))))))))))((((..-...)))---)..)))...)))))))..(((((....))))).(((((..-..)))))--------- ( -44.90, z-score = -3.14, R) >droSim1.chr3R 24247316 101 - 27517382 CGCUUGUCGGUU-GGCCAA-AGCCAGAACUGGAUUGGCUUUGCCGCCCGG-UUGGGG---CGACAAAGUGUGAGCGGAGUGCAAGUAUGCAUACUUCUGG-GGGGAAG--------- (((((.((((((-(((...-.))))..)))))....((((((.(((((..-...)))---)).))))))..)))))..(((((....))))).(((((..-..)))))--------- ( -38.40, z-score = -1.14, R) >droSec1.super_4 3430166 101 - 6179234 CGCUUGUCGGUU-GGGCAA-AGCCAGAACUGGAUUGGCUUUGCAGCCCGG-UUAGGG---CGACAAAGUGUGAGCGGAGUGCAAGUAUGCAUACUUCUGG-GGGGAAG--------- ((((..(...((-(.((((-((((((.......)))))))))).((((..-...)))---)..)))...)..))))..(((((....))))).(((((..-..)))))--------- ( -39.40, z-score = -2.42, R) >droYak2.chr3R 6891230 103 + 28832112 CGCUUGUCGGUU-GGGCAA-AGCCAGAACUGGAUUGGCUUUGCCGCCCGGUUUGGGG---CGACAAAGUGCGAGCGGAGUGCAAGUAUGCAUACUUCUGGUGGGGAAG--------- (((((((...((-((((((-((((((.......)))))))))))((((......)))---)..)))...)))))))..(((((....))))).(((((.....)))))--------- ( -45.10, z-score = -3.08, R) >droEre2.scaffold_4820 6995447 101 + 10470090 CGCUUGUCGGUC-GGGCAA-AGCCAGAACUGGAUUGGCUUUGCCGCCCGG-CUGGGG---CGACAAAGUGCGAGCGGAGUGCAAGUAUGCAUACUUAUGG-GAGGAAG--------- ((((((((.(((-.(((((-((((((.......)))))))))))((((..-...)))---))))...).)))))))..(((((....))))).(((....-)))....--------- ( -43.70, z-score = -2.85, R) >droAna3.scaffold_13340 17354262 117 - 23697760 CCCUCGUCGGCCAGAACAACAGCCAAAAAUGGAUUGGCUUGCCGACUUGAUCCGGGAAUCCGGUAAAAUGGAAGGAGUCGGGGAGUAGGAGUCCUUGCAUACUUUAAGGGUCUGGAG .....((((((.((.....((((((....))).))).)).))))))....(((((..((((..(((((((.(((((.((.........)).))))).)))..)))).))))))))). ( -37.60, z-score = -0.35, R) >consensus CGCUUGUCGGUU_GGGCAA_AGCCAGAACUGGAUUGGCUUUGCCGCCCGG_UUGGGG___CGACAAAGUGCGAGCGGAGUGCAAGUAUGCAUACUUCUGG_GGGGAAG_________ (((((((((.....(((((.((((((.......))))))))))).(((.....)))....))))).......))))..(((((....))))).(((((.....)))))......... (-22.47 = -23.98 + 1.51)
| Location | 24,548,587 – 24,548,695 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.57 |
| Shannon entropy | 0.35788 |
| G+C content | 0.55113 |
| Mean single sequence MFE | -35.18 |
| Consensus MFE | -15.08 |
| Energy contribution | -15.94 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.698507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
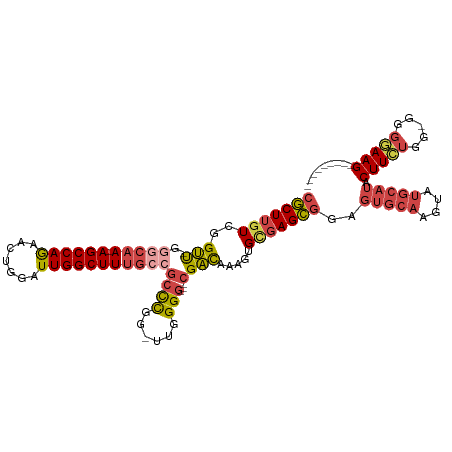
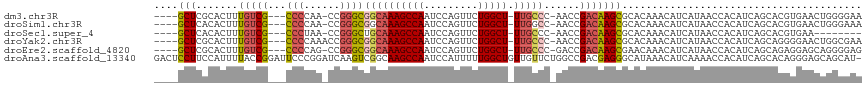
>dm3.chr3R 24548587 108 + 27905053 ----GCUCGCACUUUGUCG---CCCCAA-CCGGGCGGCAAAGCCAAUCCAGUUCUGGCU-UUGCCC-AACCGACAAGCGCACAAACAUCAUAACCACAUCAGCACGUGAACUGGGGAA ----.(((((.(((.((((---(((...-..))))((((((((((.........)))))-))))).-....)))))).))..............(((........)))....)))... ( -36.90, z-score = -2.65, R) >droSim1.chr3R 24247348 108 + 27517382 ----GCUCACACUUUGUCG---CCCCAA-CCGGGCGGCAAAGCCAAUCCAGUUCUGGCU-UUGGCC-AACCGACAAGCGCACAAACAUCAUAACCACAUCAGCACGUGAACUGGGAAA ----.......((((((((---(((...-..)))))))))))....((((((((((((.-...)))-)........((.......................))....))))))))... ( -35.80, z-score = -2.91, R) >droSec1.super_4 3430198 100 + 6179234 ----GCUCACACUUUGUCG---CCCUAA-CCGGGCUGCAAAGCCAAUCCAGUUCUGGCU-UUGCCC-AACCGACAAGCGCACAAACAUCAUAACCACAUCAGCACGUGAA-------- ----..((((...((((((---(((...-..)))).(((((((((.........)))))-))))..-....)))))((.......................))..)))).-------- ( -30.50, z-score = -3.79, R) >droYak2.chr3R 6891263 109 - 28832112 ----GCUCGCACUUUGUCG---CCCCAAACCGGGCGGCAAAGCCAAUCCAGUUCUGGCU-UUGCCC-AACCGACAAGCGCACAAACAUCAUAACCACAUCAGCAGGGGAACUGGCGAA ----..((((..(((((((---(((......))))((((((((((.........)))))-))))).-....)))))).........................(((.....))))))). ( -38.10, z-score = -2.60, R) >droEre2.scaffold_4820 6995479 108 - 10470090 ----GCUCGCACUUUGUCG---CCCCAG-CCGGGCGGCAAAGCCAAUCCAGUUCUGGCU-UUGCCC-GACCGACAAGCGAACAAACAUCAUAACCACAUCAGCAGAGGAGCAGGGGAG ----((((.....((((((---(((...-..))))((((((((((.........)))))-))))).-....)))))((.......................))....))))....... ( -36.30, z-score = -1.62, R) >droAna3.scaffold_13340 17354300 117 + 23697760 GACUCCUUCCAUUUUACCGGAUUCCCGGAUCAAGUCGGCAAGCCAAUCCAUUUUUGGCUGUUGUUCUGGCCGACGAGGGCAUAAACAUCAAAACCACAUCAGCACAGGGAGCAGCAU- (.((((((((......((((....)))).....((((((.((((((.......)))))).........))))))..))((.....................))..))))))).....- ( -33.50, z-score = -1.43, R) >consensus ____GCUCGCACUUUGUCG___CCCCAA_CCGGGCGGCAAAGCCAAUCCAGUUCUGGCU_UUGCCC_AACCGACAAGCGCACAAACAUCAUAACCACAUCAGCACGGGAACUGGGGAA ....(((.......(((((....(((.....))).((((((((((.........))))).))))).....))))))))........................................ (-15.08 = -15.94 + 0.87)
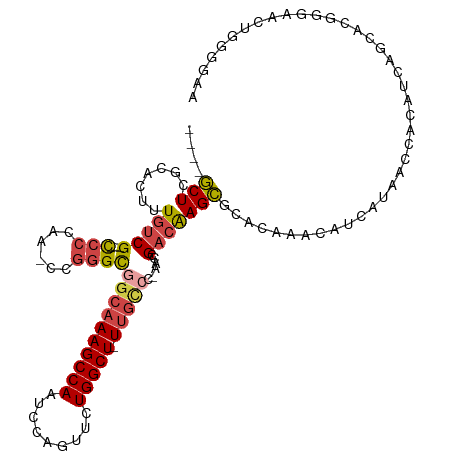
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:52:17 2011