| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,547,489 – 24,547,589 |
| Length | 100 |
| Max. P | 0.855315 |

| Location | 24,547,489 – 24,547,589 |
|---|---|
| Length | 100 |
| Sequences | 10 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 68.59 |
| Shannon entropy | 0.64017 |
| G+C content | 0.54337 |
| Mean single sequence MFE | -16.82 |
| Consensus MFE | -8.05 |
| Energy contribution | -8.56 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.855315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
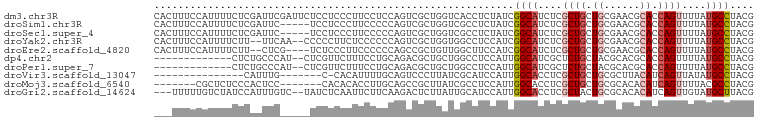
>dm3.chr3R 24547489 100 + 27905053 CACUUUCCAUUUUCUCGAUUCGAUUCUCCUCCCUUCCUCCAGUCGCUGGUCACCUCUAUCGGCAUCUCGCUGCUGCGAACGCACCAGUUUUAUGCCUACG ................(((((((((...............)))))..)))).........(((((...((((.(((....))).))))...))))).... ( -18.46, z-score = -1.86, R) >droSim1.chr3R 24246249 95 + 27517382 CACUUUCCAUUUUCUCGAUUC-----UCCUCCCUUCCCCCAGUCGCUGGUCGCCUCUAUCGGCAUCUCGCUGCUGCGAACGCACCAGUUUUAUGCCUACG .....................-----............((((...))))...........(((((...((((.(((....))).))))...))))).... ( -18.20, z-score = -1.86, R) >droSec1.super_4 3429111 95 + 6179234 CACUUUCCAUUUUCUCGAUUC-----UCCUCCCUUCCCCCAGUCGCUGGUCGCCUCUAUCGGCAUCUCGCUGCUGCGAACGCACCAGUUUUAUGCCUACG .....................-----............((((...))))...........(((((...((((.(((....))).))))...))))).... ( -18.20, z-score = -1.86, R) >droYak2.chr3R 6890119 96 - 28832112 CACUUUCCAUUUUCUU--UUCAA--CCCCCUUCUCCCCCCAGUCGCUGGUGGCCUCCAUCGGCAUCUCGCUGCUGCGAACGCACCAGUUUUAUGCCUACG ................--.....--...........((((((...)))).))........(((((...((((.(((....))).))))...))))).... ( -20.80, z-score = -2.49, R) >droEre2.scaffold_4820 6994402 94 - 10470090 CACUUUCCAUUUUCUU--CUCG----UCUCCCUUCCCCCCAGCCGCUGUUGGCUUCCAUCGGCAUCUCGCUGCUGCGAACGCACCAGUUUUAUGCCUACG ................--....----..............(((((....)))))......(((((...((((.(((....))).))))...))))).... ( -20.20, z-score = -2.60, R) >dp4.chr2 27332423 85 - 30794189 -------------CUCUGCCCAU--CUCGUUCUUUCCUGCAGACGCUGCUGGCCUCCAUUGGCAUCGCUCUGCUACGCACGCACCAGUUUUAUGCCUACG -------------.(((((....--.............))))).((.(((((((......)))...((..(((...))).))..)))).....))..... ( -16.23, z-score = 0.47, R) >droPer1.super_7 878442 85 + 4445127 -------------CUCUGCCCAU--CUCGUUCUUUCCUGCAGACGCUGCUGGCCUCCAUUGGCAUCGCUCUGCUACGCACGCACCAGUUUUAUGCCUACG -------------.(((((....--.............))))).((.(((((((......)))...((..(((...))).))..)))).....))..... ( -16.23, z-score = 0.47, R) >droVir3.scaffold_13047 8873171 77 - 19223366 ---------------CAUUUG-------C-CACAUUUUGCAGUCCCUUAUCGCAUCCAUUGGCACCUCGCUGCUGCGCUUACAUCAGUUAUAUGCCUACG ---------------....((-------(-((.....(((...........))).....)))))....((((.((......)).))))............ ( -11.60, z-score = 0.37, R) >droMoj3.scaffold_6540 5662772 86 - 34148556 -------CGCUCUCCCACUCC-------CACACACCUUGCAGCCGCUUAUCGCCUCCAUUGGCACCUCGCUGCUGCGCACACAUCAGUUUUACGCCUACG -------..............-------..........(((((.((.....(((......))).....)).)))))........................ ( -16.50, z-score = -1.63, R) >droGri2.scaffold_14624 4156931 95 - 4233967 ---UUUUUGUCUAUCCAUUUGUC--UAUCUCAAUUCUUCAAGACUCUUAUUGCAUCCAUUGGCACCUCGCUACUGCGCACACAUCAGUUGUAUGCUUACG ---.....((((......(((..--.....))).......)))).......(((.....((((.....)))).)))(((.(((.....))).)))..... ( -11.82, z-score = -0.37, R) >consensus ___UUUCCAUUUUCUCAAUUC_____UCCUCCCUUCCUCCAGUCGCUGGUCGCCUCCAUCGGCAUCUCGCUGCUGCGAACGCACCAGUUUUAUGCCUACG ............................................................((((....((((.((......)).))))....)))).... ( -8.05 = -8.56 + 0.51)

| Location | 24,547,489 – 24,547,589 |
|---|---|
| Length | 100 |
| Sequences | 10 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 68.59 |
| Shannon entropy | 0.64017 |
| G+C content | 0.54337 |
| Mean single sequence MFE | -22.42 |
| Consensus MFE | -15.97 |
| Energy contribution | -15.43 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.53 |
| Mean z-score | -0.31 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.833548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
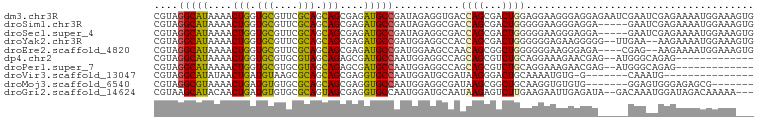
>dm3.chr3R 24547489 100 - 27905053 CGUAGGCAUAAAACUGGUGCGUUCGCAGCAGCGAGAUGCCGAUAGAGGUGACCAGCGACUGGAGGAAGGGAGGAGAAUCGAAUCGAGAAAAUGGAAAGUG ....(((((....(((.(((....))).)))....)))))...........(((....((.((....((........))...)).))....)))...... ( -23.20, z-score = -0.59, R) >droSim1.chr3R 24246249 95 - 27517382 CGUAGGCAUAAAACUGGUGCGUUCGCAGCAGCGAGAUGCCGAUAGAGGCGACCAGCGACUGGGGGAAGGGAGGA-----GAAUCGAGAAAAUGGAAAGUG (((.(((((....(((.(((....))).)))....))))).......))).(((....((.((...........-----...)).))....)))...... ( -21.04, z-score = 0.15, R) >droSec1.super_4 3429111 95 - 6179234 CGUAGGCAUAAAACUGGUGCGUUCGCAGCAGCGAGAUGCCGAUAGAGGCGACCAGCGACUGGGGGAAGGGAGGA-----GAAUCGAGAAAAUGGAAAGUG (((.(((((....(((.(((....))).)))....))))).......))).(((....((.((...........-----...)).))....)))...... ( -21.04, z-score = 0.15, R) >droYak2.chr3R 6890119 96 + 28832112 CGUAGGCAUAAAACUGGUGCGUUCGCAGCAGCGAGAUGCCGAUGGAGGCCACCAGCGACUGGGGGGAGAAGGGGG--UUGAA--AAGAAAAUGGAAAGUG (((.(((((....(((.(((....))).)))....))))).)))..((((.((..(..(.....)..)...))))--))...--................ ( -25.50, z-score = -0.81, R) >droEre2.scaffold_4820 6994402 94 + 10470090 CGUAGGCAUAAAACUGGUGCGUUCGCAGCAGCGAGAUGCCGAUGGAAGCCAACAGCGGCUGGGGGGAAGGGAGA----CGAG--AAGAAAAUGGAAAGUG (((.(((((....(((.(((....))).)))....))))).)))..((((......)))).........(....----)...--................ ( -25.00, z-score = -1.22, R) >dp4.chr2 27332423 85 + 30794189 CGUAGGCAUAAAACUGGUGCGUGCGUAGCAGAGCGAUGCCAAUGGAGGCCAGCAGCGUCUGCAGGAAAGAACGAG--AUGGGCAGAG------------- (((.(((((....(((.(((....))).)))....))))).)))..(.(((....(((((.......)).)))..--.))).)....------------- ( -25.10, z-score = -0.59, R) >droPer1.super_7 878442 85 - 4445127 CGUAGGCAUAAAACUGGUGCGUGCGUAGCAGAGCGAUGCCAAUGGAGGCCAGCAGCGUCUGCAGGAAAGAACGAG--AUGGGCAGAG------------- (((.(((((....(((.(((....))).)))....))))).)))..(.(((....(((((.......)).)))..--.))).)....------------- ( -25.10, z-score = -0.59, R) >droVir3.scaffold_13047 8873171 77 + 19223366 CGUAGGCAUAUAACUGAUGUAAGCGCAGCAGCGAGGUGCCAAUGGAUGCGAUAAGGGACUGCAAAAUGUG-G-------CAAAUG--------------- (((.(((((....(((.(((....))).)))....))))).)))..(((.(((.............))).-)-------))....--------------- ( -16.72, z-score = 0.67, R) >droMoj3.scaffold_6540 5662772 86 + 34148556 CGUAGGCGUAAAACUGAUGUGUGCGCAGCAGCGAGGUGCCAAUGGAGGCGAUAAGCGGCUGCAAGGUGUGUG-------GGAGUGGGAGAGCG------- .(((.((((.......)))).)))(((((.((....((((......))))....)).)))))..........-------..............------- ( -25.30, z-score = -0.34, R) >droGri2.scaffold_14624 4156931 95 + 4233967 CGUAAGCAUACAACUGAUGUGUGCGCAGUAGCGAGGUGCCAAUGGAUGCAAUAAGAGUCUUGAAGAAUUGAGAUA--GACAAAUGGAUAGACAAAAA--- .(((.(((((((.....)))))))(((.(.....).))).......))).......((((..(....)..)))).--....................--- ( -16.20, z-score = 0.06, R) >consensus CGUAGGCAUAAAACUGGUGCGUGCGCAGCAGCGAGAUGCCAAUGGAGGCGACCAGCGACUGGAGGAAGGGAGGA_____GAAUCGAGAAAAUGGAAA___ ....(((((....(((.(((....))).)))....)))))...........((((...))))...................................... (-15.97 = -15.43 + -0.54)
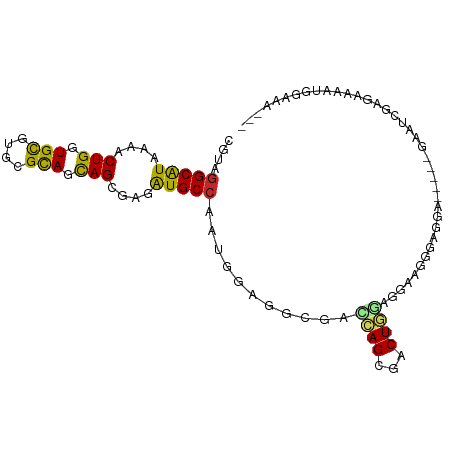
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:52:15 2011