| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,542,626 – 24,542,751 |
| Length | 125 |
| Max. P | 0.973350 |

| Location | 24,542,626 – 24,542,751 |
|---|---|
| Length | 125 |
| Sequences | 6 |
| Columns | 132 |
| Reading direction | forward |
| Mean pairwise identity | 81.91 |
| Shannon entropy | 0.34538 |
| G+C content | 0.48648 |
| Mean single sequence MFE | -38.62 |
| Consensus MFE | -17.57 |
| Energy contribution | -18.58 |
| Covariance contribution | 1.02 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.827109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
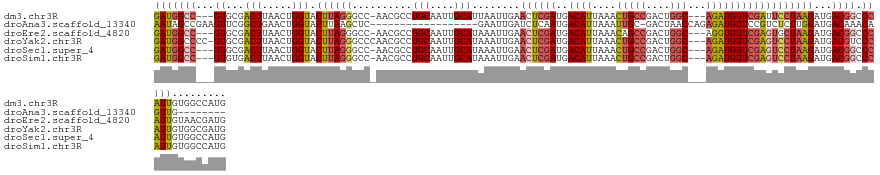
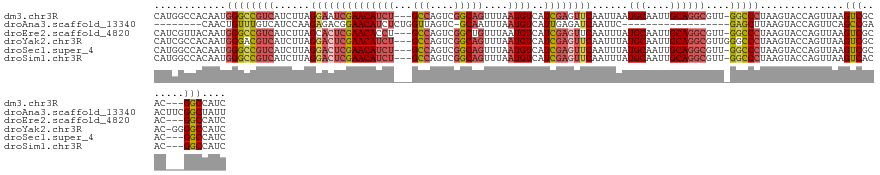
>dm3.chr3R 24542626 125 + 27905053 GAUGGCC---GUGCGACUUAACUGGUACUUAGGGCC-AACGCCUGCAAUUGCAUUAAUUGAACUCGAUGACAUUAAACUGCCGACUGGC---AGAUGUUCGAUUCCUAAGAUGACGGCCCAUUGUGGCCAUG .((((((---((((.(......).))))...(((((-......(((....)))......(((.((((..((((....(((((....)))---)))))))))))))..........))))).....)))))). ( -40.90, z-score = -3.08, R) >droAna3.scaffold_13340 17348436 105 + 23697760 AAUAGCCGAAGUUCGGCUGAACUGGUACUUAAGCUC------------------GAAUUGAUCUCAAUGACAUUAAAUUGC-GACUAACCAGAGAUGUUCCGUCUCUUGGAUGACAAACAGUUG-------- ..((((((.....))))))(((((............------------------...(((.((.((((........)))).-)).)))(((((((((...)))))).)))........))))).-------- ( -26.50, z-score = -1.65, R) >droEre2.scaffold_4820 6989536 125 - 10470090 GAUGGCC---GUGCGACUUAACUGGUACUUAGGGCC-AACGCCUGCAAUUGCAUAAAUUGAACUCGAUGACAUUAAACAGCCGACUGGC---AGGUGUUCGAGUGCUAAGAUGACGGCCCAUUGUAACGAUG ...((((---((.(((((.....))).(((((.(((-(((((((((..........((((....)))).........(((....)))))---))))))).).)).))))).))))))))(((((...))))) ( -38.60, z-score = -1.94, R) >droYak2.chr3R 6885212 128 - 28832112 GAUGGCCCC-GUGCGACUUAACUGGUACUUAGGGCCCAACGCCUGCAAUUGCAUAAAUUGAACUCGAUGACAUUAAACUGCCGACUGGC---AGAUGUUCGAGUCCUAAGAUGACGUCCCAUUGUGGCGAUG ...(((((.-((((.(......).))))...)))))...((((.((((((((((.....(.((((((..((((....(((((....)))---)))))))))))).)....))).))....)))))))))... ( -39.90, z-score = -2.56, R) >droSec1.super_4 3424266 125 + 6179234 GAUGGCC---GUGCGACUUAACUGGUACUUAGGGCC-AACGCCUGCAAUUGCAUAAAUUGAACUCGAUGACAUUAAACUGCCGACUGGC---AGAUGUUCGAGUCCUAAGAUGACGGCCCAUUGUGGCCAUG .((((((---((((.(......).))))...(((((-..((.((.(((((.....))))).((((((..((((....(((((....)))---))))))))))))....)).))..))))).....)))))). ( -44.20, z-score = -3.72, R) >droSim1.chr3R 24240926 125 + 27517382 GAUGGCC---GUGUGACUUAACUGGUACUUAGGGCC-AACGCCUGCAAUUGCAUAAAUUGAACUCGAUGACAUUAAACUGCCGACUGGC---AGAUGUUCGAGUCCUAAGAUGACGGCCCAUUGUGGCCAUG .((((((---(.(((.((.....)))))...(((((-..((.((.(((((.....))))).((((((..((((....(((((....)))---))))))))))))....)).))..)))))....))))))). ( -41.60, z-score = -2.99, R) >consensus GAUGGCC___GUGCGACUUAACUGGUACUUAGGGCC_AACGCCUGCAAUUGCAUAAAUUGAACUCGAUGACAUUAAACUGCCGACUGGC___AGAUGUUCGAGUCCUAAGAUGACGGCCCAUUGUGGCCAUG (((((..........(((.....))).((((((..........(((....)))........((((((..(((((.....(((....)))....)))))))))))))))))........)))))......... (-17.57 = -18.58 + 1.02)
| Location | 24,542,626 – 24,542,751 |
|---|---|
| Length | 125 |
| Sequences | 6 |
| Columns | 132 |
| Reading direction | reverse |
| Mean pairwise identity | 81.91 |
| Shannon entropy | 0.34538 |
| G+C content | 0.48648 |
| Mean single sequence MFE | -39.15 |
| Consensus MFE | -22.22 |
| Energy contribution | -22.85 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
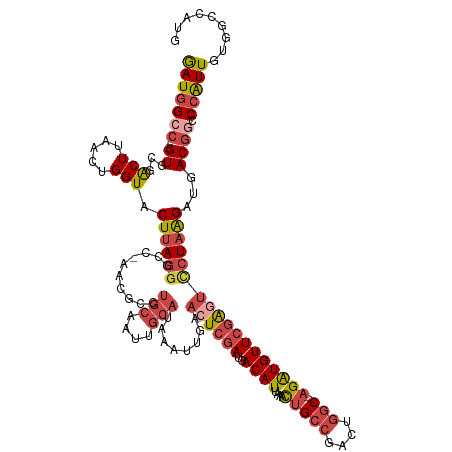
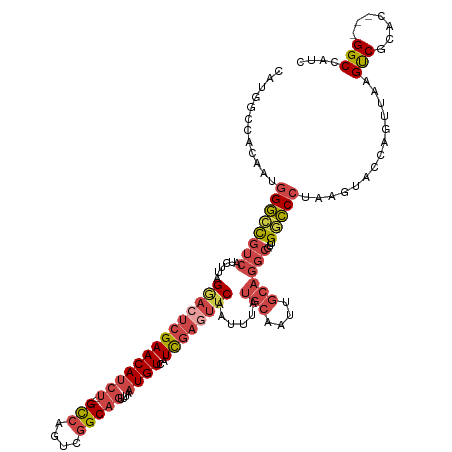
>dm3.chr3R 24542626 125 - 27905053 CAUGGCCACAAUGGGCCGUCAUCUUAGGAAUCGAACAUCU---GCCAGUCGGCAGUUUAAUGUCAUCGAGUUCAAUUAAUGCAAUUGCAGGCGUU-GGCCCUAAGUACCAGUUAAGUCGCAC---GGCCAUC .((((((.....((((((((......(((.((((((((((---(((....)))))....))))..)))).)))......(((....))))))...-)))))...((..(......)..))..---)))))). ( -39.50, z-score = -2.35, R) >droAna3.scaffold_13340 17348436 105 - 23697760 --------CAACUGUUUGUCAUCCAAGAGACGGAACAUCUCUGGUUAGUC-GCAAUUUAAUGUCAUUGAGAUCAAUUC------------------GAGCUUAAGUACCAGUUCAGCCGAACUUCGGCUAUU --------.(((((((((((........))))))....(((..(((.(((-.((((........)))).))).)))..------------------))).........))))).(((((.....)))))... ( -23.70, z-score = -0.80, R) >droEre2.scaffold_4820 6989536 125 + 10470090 CAUCGUUACAAUGGGCCGUCAUCUUAGCACUCGAACACCU---GCCAGUCGGCUGUUUAAUGUCAUCGAGUUCAAUUUAUGCAAUUGCAGGCGUU-GGCCCUAAGUACCAGUUAAGUCGCAC---GGCCAUC .............(((((((..(((((((((((((((...---(((....))).......)))..))))))...((((((((....)))(((...-.))).)))))....))))))..).))---))))... ( -35.90, z-score = -1.69, R) >droYak2.chr3R 6885212 128 + 28832112 CAUCGCCACAAUGGGACGUCAUCUUAGGACUCGAACAUCU---GCCAGUCGGCAGUUUAAUGUCAUCGAGUUCAAUUUAUGCAAUUGCAGGCGUUGGGCCCUAAGUACCAGUUAAGUCGCAC-GGGGCCAUC ((.((((....(((((.....)))))((((((((((((((---(((....)))))....))))..))))))))......(((....))))))).))((((((..((..(......)..))..-))))))... ( -45.10, z-score = -3.50, R) >droSec1.super_4 3424266 125 - 6179234 CAUGGCCACAAUGGGCCGUCAUCUUAGGACUCGAACAUCU---GCCAGUCGGCAGUUUAAUGUCAUCGAGUUCAAUUUAUGCAAUUGCAGGCGUU-GGCCCUAAGUACCAGUUAAGUCGCAC---GGCCAUC .((((((.....((((((((......((((((((((((((---(((....)))))....))))..))))))))......(((....))))))...-)))))...((..(......)..))..---)))))). ( -45.50, z-score = -3.94, R) >droSim1.chr3R 24240926 125 - 27517382 CAUGGCCACAAUGGGCCGUCAUCUUAGGACUCGAACAUCU---GCCAGUCGGCAGUUUAAUGUCAUCGAGUUCAAUUUAUGCAAUUGCAGGCGUU-GGCCCUAAGUACCAGUUAAGUCACAC---GGCCAUC .((((((.....((((((((......((((((((((((((---(((....)))))....))))..))))))))......(((....))))))...-)))))...((..(......)..))..---)))))). ( -45.20, z-score = -4.23, R) >consensus CAUGGCCACAAUGGGCCGUCAUCUUAGGACUCGAACAUCU___GCCAGUCGGCAGUUUAAUGUCAUCGAGUUCAAUUUAUGCAAUUGCAGGCGUU_GGCCCUAAGUACCAGUUAAGUCGCAC___GGCCAUC ............(((((.........((((((((((((.....(((....)))......))))..))))))))......(((....))).......)))))..............(((.......))).... (-22.22 = -22.85 + 0.63)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:52:13 2011