| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,522,374 – 24,522,465 |
| Length | 91 |
| Max. P | 0.781539 |

| Location | 24,522,374 – 24,522,465 |
|---|---|
| Length | 91 |
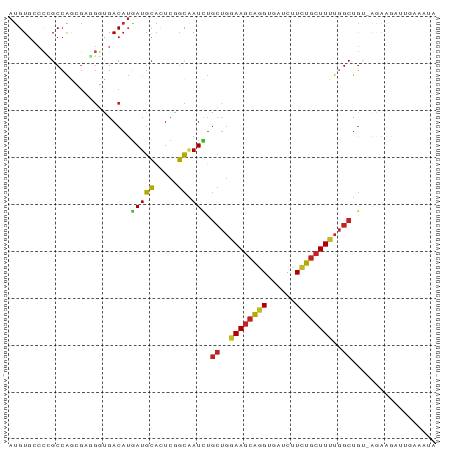
| Sequences | 11 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 72.14 |
| Shannon entropy | 0.62664 |
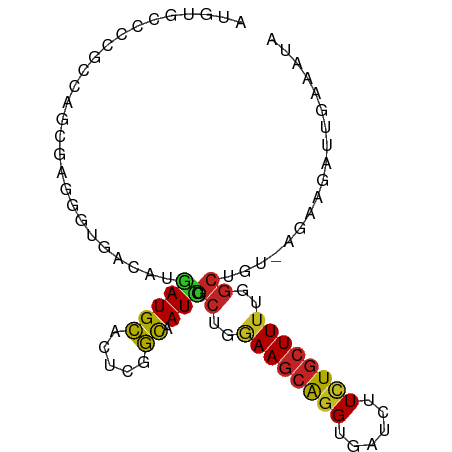
| G+C content | 0.50949 |
| Mean single sequence MFE | -29.01 |
| Consensus MFE | -15.12 |
| Energy contribution | -14.83 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.47 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.781539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24522374 91 - 27905053 AUGUGGCCCGCCAGCGAGGGCGACAUGAUGCACUCGGCAAUCUGCUGAAAGCAGGUGAUCUUCUGCUUUUGGCUGUGGGGGGUUCGAAAUA ((((.((((........)))).))))((.((.(((.(((....((..((((((((......))))))))..))))).))).))))...... ( -36.90, z-score = -2.01, R) >anoGam1.chr2R 43847384 71 - 62725911 ACGUGCCGGGCCGA--ACGUUGAC--GAUGGUUUCACUGAUCACACCGAAAUGGGCCACCUUUUCCUUGCCGCUG---------------- ........((((.(--...(((..--(.(((((.....))))))..)))..).))))..................---------------- ( -11.60, z-score = 2.45, R) >droEre2.scaffold_4820 6965475 90 + 10470090 AUGUGCCCCGCCAGCGAGGGCGACAUGAUGCACUCGGCAAUCUGCUGGAAGCAGGUGAUCUUCUGCUUUUGGCUGU-GGGAAUUUGAGUUA ((((((((((....)).)))).)))).....((((((...((.((..((((((((......))))))))..))...-))....)))))).. ( -34.30, z-score = -1.44, R) >droYak2.chr3R 6864529 90 + 28832112 AUGUGCCCCGCCAGCGAGGGCGACAUGAUGCACUCGGCAAUCUGCUGGAAGCAGGUGAUCUUCUGCUUUUGGCUGU-GGGAAAUUGAGGUA ((((((((((....)).)))).))))..(((.(((((...((.((..((((((((......))))))))..))...-))....)))))))) ( -35.00, z-score = -1.45, R) >droSec1.super_4 3404178 90 - 6179234 AUGUGGCCCGCCAGCGAGGGCGACAUGAUGCACUCGGCAAUCUGCUGAAAGCAGGUGAUCUUCUGCUUUUGGCUGU-GGAGAUUCGAAAUA ((((.((((........)))).))))......(((.(((....((..((((((((......))))))))..)))))-.))).......... ( -36.40, z-score = -2.47, R) >dp4.chr2 27305474 91 + 30794189 AUGUGCCCCGCCAGUGCAGGUGACAUGAUGCACUCUGCUAUCUGCUGGAAGCAGGUGAUCUUCUGCUUUUGGCUGUGAAAAGAUUUGGAAG ..........((((.(((((((.(.....)))).))))..((.((..((((((((......))))))))..))...))......))))... ( -29.10, z-score = -0.96, R) >droPer1.super_7 851591 91 - 4445127 AUGUGCCCCGCCAGUGCAGGUGACAUGAUGCACUCUGCUAUCUGCUGGAAGCAGGUGAUCUUCUGCUUUUGGCUGUGAAAAGAUUUGGAAG ..........((((.(((((((.(.....)))).))))..((.((..((((((((......))))))))..))...))......))))... ( -29.10, z-score = -0.96, R) >droAna3.scaffold_13340 17328889 90 - 23697760 AUGUGACCCGCAAGUGAAGGGGACAUUAUGCAUUCUGCAAUCUGCUGAAAGCAGGCAAUCUUUUGCUUUUGGCUGCAAAAAAUGUAAAAA- ((((..(((.........))).))))..((((((.((((....((..((((((((......))))))))..))))))...))))))....- ( -25.60, z-score = -1.26, R) >droVir3.scaffold_13047 8841963 91 + 19223366 AUGUGACCGGCCAAAGCUGCUGACAUGAUGCACUCGGCAAUCUGCUGGAAGCAUGCAAUCUUUUGCUUUUGGCUAAAAAGAGUUUACCAUA ..((.(.((((....)))).).))(((.((.((((........((..((((((..........))))))..))......)))).)).))). ( -23.34, z-score = -0.17, R) >droMoj3.scaffold_6540 5632331 88 + 34148556 AUGUGGCUGGCCAAAGCCUUUGACAUAAUGCAUUCAGCAAUUUGCUGAAAGCAGGCAAUCUUUUGCUUUUGGCUGCAAGGAUAGAAGA--- .....((.(((((((.............(((.((((((.....)))))).)))(((((....))))))))))))))............--- ( -27.30, z-score = -0.63, R) >droGri2.scaffold_14624 4122448 87 + 4233967 AUGUGAGCGGCCAGCGCCGGUGACAUGAUGCACUCGGCAAUUUGCUGGAAGCAGGCAAUCUUUUGCUUUUGGCUGUGAGAUGAGACA---- ((((.(.((((....)))).).))))......((((.((....((..((((((((......))))))))..))..))...))))...---- ( -30.50, z-score = -0.88, R) >consensus AUGUGCCCCGCCAGCGAGGGUGACAUGAUGCACUCGGCAAUCUGCUGGAAGCAGGUGAUCUUCUGCUUUUGGCUGU_AGAAGAUUGAAAUA ..........................(((((.....)).))).((..((((((((......))))))))..)).................. (-15.12 = -14.83 + -0.29)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:52:05 2011