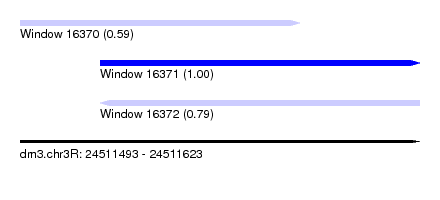
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,511,493 – 24,511,623 |
| Length | 130 |
| Max. P | 0.999320 |

| Location | 24,511,493 – 24,511,584 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 67.54 |
| Shannon entropy | 0.59240 |
| G+C content | 0.40744 |
| Mean single sequence MFE | -22.73 |
| Consensus MFE | -7.77 |
| Energy contribution | -7.60 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.587807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
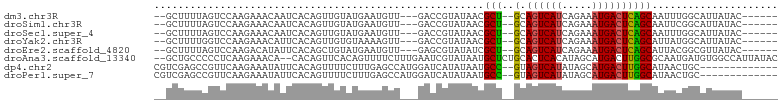
>dm3.chr3R 24511493 91 + 27905053 --GCUUUUAGUCCAAGAAACAAUCACAGUUGUAUGAAUGUU---GACCGUAUAACGCU--GCAGUCAUCAGAAAUGACUCAGCAAUUUGGCAUUAUAC------ --.....(((((((((...........((((((((......---...))))))))(((--(.((((((.....))))))))))..))))).))))...------ ( -24.40, z-score = -2.69, R) >droSim1.chr3R 24208897 91 + 27517382 --GCUUUUAGUCCAAGAAACAAUCACAGUUGUAUGAAUGUU---GACCGUAUAACGCU--GCAGUCAUCAGAAAUGACUCAGCAAUUCGGCAUUAUAC------ --.....((((((..............((((((((......---...))))))))(((--(.((((((.....)))))))))).....)).))))...------ ( -22.20, z-score = -2.07, R) >droSec1.super_4 3393514 91 + 6179234 --GCUUUUAGUCCAAGAAACAAUCACAGUUGUAUGAAUGUU---GACCGUAUAACGCU--GCAGUCAUCAGAAAUGACUCAGCAAUUUGGCAUUAUAC------ --.....(((((((((...........((((((((......---...))))))))(((--(.((((((.....))))))))))..))))).))))...------ ( -24.40, z-score = -2.69, R) >droYak2.chr3R 6853554 91 - 28832112 --GCUUUUGGUCCAAGAAACAUUCACAGUUGUGUAAAAGUU---GACCGUAUAACGCU--GCAGUCAUCAGAAAUGACUCAGCAUUAUGGCAUUAUAC------ --((....((((......((((........)))).......---)))).(((((.(((--(.((((((.....)))))))))).))))))).......------ ( -22.62, z-score = -1.75, R) >droEre2.scaffold_4820 6954410 91 - 10470090 --GCUUUUAGUCCAAGACAUAUUCACAGCUGUAUGAAUGUU---GAGCGUAUAUCGCU--GCAGUCAUCAGAAAUGACUCAGCAUUACGGCGUUAUAC------ --.......(((...))).........((((((...(((((---(((((.....))))--..((((((.....)))))))))))))))))).......------ ( -22.70, z-score = -1.06, R) >droAna3.scaffold_13340 17317999 100 + 23697760 --GCUGCCCCCUCAAGAAACA--CACAGUUCACAGUUUUCUUUGAAUCGUAUAAUGCUCUGCACUCACAUAGCAUGACUUGGCGCAAUGAUGUGGCCAUUAUAC --((.(((...((........--.((.(((((.((....)).))))).))...(((((.((......)).)))))))...))))).((((((....)))))).. ( -19.70, z-score = 0.08, R) >dp4.chr2 27290048 89 - 30794189 CGUCGAGCCGUUCAAGAAAUAUUCACAGUUUUCUUUGAGCCAUGGAUCAUAUAAUGCC--GUAGUCAUAUAGCAUGACUUGGCAUAACUGC------------- ....((.((((((((((((..........)))).))))))...)).)).....(((((--(.((((((.....))))))))))))......------------- ( -22.90, z-score = -1.88, R) >droPer1.super_7 840484 89 + 4445127 CGUCGAGCCGUUCAAGAAAUAUUCACAGUUUUCUUUGAGCCAUGGAUCAUAUAAUGCC--GUAGUCAUAUAGCAUGACUUGGCAUAACUGC------------- ....((.((((((((((((..........)))).))))))...)).)).....(((((--(.((((((.....))))))))))))......------------- ( -22.90, z-score = -1.88, R) >consensus __GCUUUUAGUCCAAGAAACAUUCACAGUUGUAUGAAUGUU___GACCGUAUAACGCU__GCAGUCAUCAGAAAUGACUCAGCAUUACGGCAUUAUAC______ .......................................................(((....((((((.....)))))).)))..................... ( -7.77 = -7.60 + -0.17)

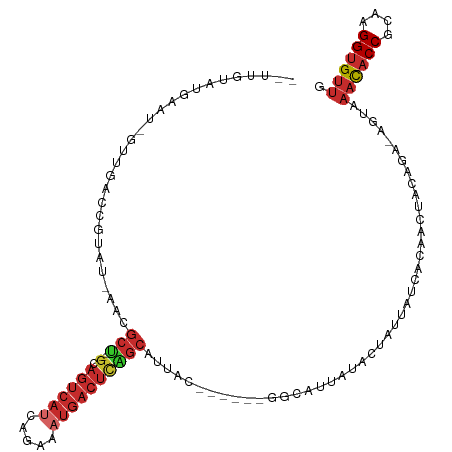
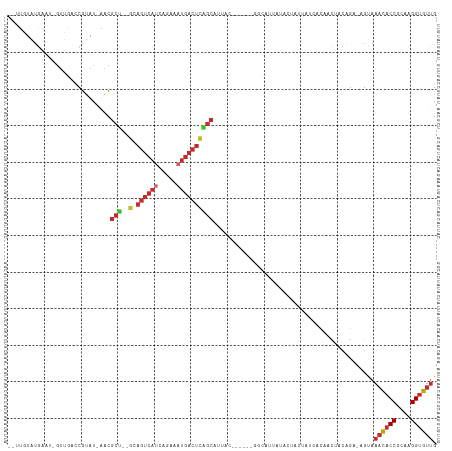
| Location | 24,511,519 – 24,511,623 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 67.02 |
| Shannon entropy | 0.64048 |
| G+C content | 0.40073 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -14.05 |
| Energy contribution | -15.07 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.79 |
| SVM RNA-class probability | 0.999320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
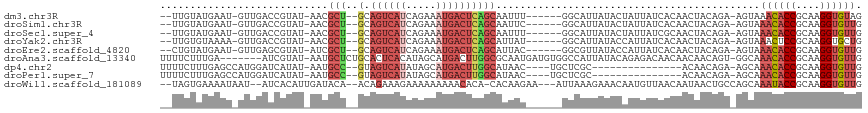
>dm3.chr3R 24511519 104 + 27905053 --UUGUAUGAAU-GUUGACCGUAU-AACGCU--GCAGUCAUCAGAAAUGACUCAGCAAUUU------GGCAUUAUACUAUUAUCACAACUACAGA-AGUAAACACCGCAAGGUGUAG --..((((((.(-((..(..(...-..)(((--(.((((((.....))))))))))...).------.)))))))))..................-.....(((((....))))).. ( -29.40, z-score = -3.14, R) >droSim1.chr3R 24208923 104 + 27517382 --UUGUAUGAAU-GUUGACCGUAU-AACGCU--GCAGUCAUCAGAAAUGACUCAGCAAUUC------GGCAUUAUACUAUUAUCACAACUACAGA-AGUAAACACCGCAAGGUGUUG --..((((((.(-(((((..(...-..)(((--(.((((((.....))))))))))...))------))))))))))..................-....((((((....)))))). ( -31.70, z-score = -3.83, R) >droSec1.super_4 3393540 104 + 6179234 --UUGUAUGAAU-GUUGACCGUAU-AACGCU--GCAGUCAUCAGAAAUGACUCAGCAAUUU------GGCAUUAUACUAUUAUCGCAACUACAGA-AGUAAACACCGCAAGGUGUUG --..((((((.(-((..(..(...-..)(((--(.((((((.....))))))))))...).------.)))))))))..................-....((((((....)))))). ( -30.30, z-score = -2.99, R) >droYak2.chr3R 6853580 104 - 28832112 --UUGUGUAAAA-GUUGACCGUAU-AACGCU--GCAGUCAUCAGAAAUGACUCAGCAUUAU------GGCAUUAUACCAUUAUCACAACUACAGA-AGUAAACUCCGCAAGGUGCUG --((((((((..-((((.(((((.-...(((--(.((((((.....))))))))))..)))------))))....))..))).))))).......-((((....((....)))))). ( -25.30, z-score = -1.69, R) >droEre2.scaffold_4820 6954436 104 - 10470090 --CUGUAUGAAU-GUUGAGCGUAU-AUCGCU--GCAGUCAUCAGAAAUGACUCAGCAUUAC------GGCGUUAUACCAUUAUCACAACUACAGA-AGUAAACACCGCAAGGUGUUG --((((((((((-(...(((((..-...(((--(.((((((.....)))))))))).....------.)))))....)))..)))....))))).-....((((((....)))))). ( -32.60, z-score = -3.30, R) >droAna3.scaffold_13340 17318030 108 + 23697760 UUUUCUUUGA-------AUCGUAU-AAUGCUCUGCACUCACAUAGCAUGACUUGGCGCAAUGAUGUGGCCAUUAUACAGAGACAACAACAACAGU-GGCAAACACCGCAAGGUGUUG ...((((((.-------...((..-.(((((.((......)).))))).)).(((((((....))).)))).....)))))).............-....((((((....)))))). ( -29.10, z-score = -1.28, R) >dp4.chr2 27290076 94 - 30794189 UUUUCUUUGAGCCAUGGAUCAUAU-AAUGCC--GUAGUCAUAUAGCAUGACUUGGCAUAAC----UGCUCGC---------------ACAACAGA-AGCAAACACCGCAAGGUGUUG .((((((((.((....((.((...-.(((((--(.((((((.....))))))))))))...----)).))))---------------.))).)))-))..((((((....)))))). ( -29.20, z-score = -2.90, R) >droPer1.super_7 840512 94 + 4445127 UUUUCUUUGAGCCAUGGAUCAUAU-AAUGCC--GUAGUCAUAUAGCAUGACUUGGCAUAAC----UGCUCGC---------------ACAACAGA-AGCAAACACCGCAAGGUGUUG .((((((((.((....((.((...-.(((((--(.((((((.....))))))))))))...----)).))))---------------.))).)))-))..((((((....)))))). ( -29.20, z-score = -2.90, R) >droWil1.scaffold_181089 9224116 107 + 12369635 --UAGUGAAAAUAAU--AUCACAUUGAUACA--ACAGAAAGAAAAAAAAACACA-CACAAGAA---AUUAAAGAAACAAUGUUAACAAUAACUGCCAGCAAAUACCGCAAGGUGUUG --..((((.......--.)))).........--.....................-........---.............((((..((.....))..))))((((((....)))))). ( -13.10, z-score = -1.69, R) >consensus __UUGUAUGAAU_GUUGACCGUAU_AACGCU__GCAGUCAUCAGAAAUGACUCAGCAUUAC______GGCAUUAUACUAUUAUCACAACUACAGA_AGUAAACACCGCAAGGUGUUG ............................(((....((((((.....)))))).)))............................................((((((....)))))). (-14.05 = -15.07 + 1.01)
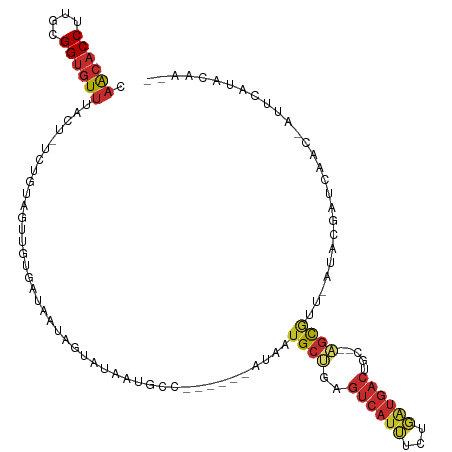
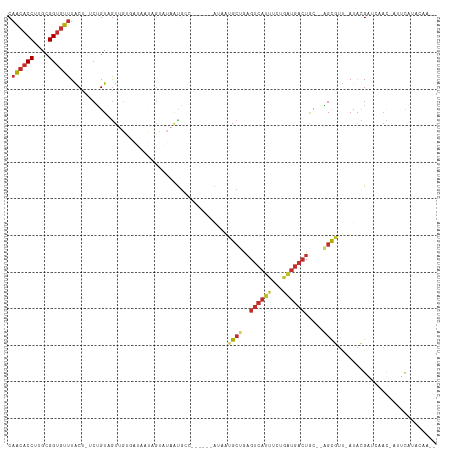
| Location | 24,511,519 – 24,511,623 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 67.02 |
| Shannon entropy | 0.64048 |
| G+C content | 0.40073 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -10.35 |
| Energy contribution | -10.82 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.791941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24511519 104 - 27905053 CUACACCUUGCGGUGUUUACU-UCUGUAGUUGUGAUAAUAGUAUAAUGCC------AAAUUGCUGAGUCAUUUCUGAUGACUGC--AGCGUU-AUACGGUCAAC-AUUCAUACAA-- ..(((((....))))).....-..((((..((((((....(((((((((.------....(((..(((((((...)))))))))--))))))-)))).))).))-)....)))).-- ( -29.10, z-score = -2.32, R) >droSim1.chr3R 24208923 104 - 27517382 CAACACCUUGCGGUGUUUACU-UCUGUAGUUGUGAUAAUAGUAUAAUGCC------GAAUUGCUGAGUCAUUUCUGAUGACUGC--AGCGUU-AUACGGUCAAC-AUUCAUACAA-- .((((((....))))))....-..((((..((((((....(((((((((.------....(((..(((((((...)))))))))--))))))-)))).))).))-)....)))).-- ( -30.10, z-score = -2.39, R) >droSec1.super_4 3393540 104 - 6179234 CAACACCUUGCGGUGUUUACU-UCUGUAGUUGCGAUAAUAGUAUAAUGCC------AAAUUGCUGAGUCAUUUCUGAUGACUGC--AGCGUU-AUACGGUCAAC-AUUCAUACAA-- .((((((....))))))....-..((((..((.(((....(((((((((.------....(((..(((((((...)))))))))--))))))-)))).)))..)-)....)))).-- ( -27.30, z-score = -1.69, R) >droYak2.chr3R 6853580 104 + 28832112 CAGCACCUUGCGGAGUUUACU-UCUGUAGUUGUGAUAAUGGUAUAAUGCC------AUAAUGCUGAGUCAUUUCUGAUGACUGC--AGCGUU-AUACGGUCAAC-UUUUACACAA-- ..((.....))((((....))-))(((((..(((((...(((.....)))------((((((((((((((((...))))))).)--))))))-))...))).))-..)))))...-- ( -30.90, z-score = -2.37, R) >droEre2.scaffold_4820 6954436 104 + 10470090 CAACACCUUGCGGUGUUUACU-UCUGUAGUUGUGAUAAUGGUAUAACGCC------GUAAUGCUGAGUCAUUUCUGAUGACUGC--AGCGAU-AUACGCUCAAC-AUUCAUACAG-- .((((((....))))))....-.(((((...(((...(((((.....)))------))..((((((((((((...))))))).)--))))..-..........)-))...)))))-- ( -30.10, z-score = -2.17, R) >droAna3.scaffold_13340 17318030 108 - 23697760 CAACACCUUGCGGUGUUUGCC-ACUGUUGUUGUUGUCUCUGUAUAAUGGCCACAUCAUUGCGCCAAGUCAUGCUAUGUGAGUGCAGAGCAUU-AUACGAU-------UCAAAGAAAA .((((((....))))))....-...(((((...((.((((((((..((((((......)).))))..((((.....))))))))))))))..-..)))))-------.......... ( -28.30, z-score = -0.31, R) >dp4.chr2 27290076 94 + 30794189 CAACACCUUGCGGUGUUUGCU-UCUGUUGU---------------GCGAGCA----GUUAUGCCAAGUCAUGCUAUAUGACUAC--GGCAUU-AUAUGAUCCAUGGCUCAAAGAAAA .((((((....))))))..((-(..((..(---------------(.((.((----...(((((.(((((((...)))))))..--))))).-...)).))))..))...))).... ( -27.70, z-score = -1.60, R) >droPer1.super_7 840512 94 - 4445127 CAACACCUUGCGGUGUUUGCU-UCUGUUGU---------------GCGAGCA----GUUAUGCCAAGUCAUGCUAUAUGACUAC--GGCAUU-AUAUGAUCCAUGGCUCAAAGAAAA .((((((....))))))..((-(..((..(---------------(.((.((----...(((((.(((((((...)))))))..--))))).-...)).))))..))...))).... ( -27.70, z-score = -1.60, R) >droWil1.scaffold_181089 9224116 107 - 12369635 CAACACCUUGCGGUAUUUGCUGGCAGUUAUUGUUAACAUUGUUUCUUUAAU---UUCUUGUG-UGUGUUUUUUUUUCUUUCUGU--UGUAUCAAUGUGAU--AUUAUUUUCACUA-- .(((((..((((((....))).)))((((....))))..............---........-.)))))...............--.........((((.--.......))))..-- ( -11.80, z-score = 0.55, R) >consensus CAACACCUUGCGGUGUUUACU_UCUGUAGUUGUGAUAAUAGUAUAAUGCC______AUAAUGCUGAGUCAUUUCUGAUGACUGC__AGCGUU_AUACGAUCAAC_AUUCAUACAA__ .((((((....))))))...........................................((((.(((((((...)))))))....))))........................... (-10.35 = -10.82 + 0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:52:03 2011